Structural and functional characterization of a highly specific serpin in the insect innate immunity
Park, S.H., Jiang, R., Piao, S., Zhang, B., Kim, E.H., Kwon, H.M., Jin, X.L., Lee, B.L., Ha, N.-C.(2011) J Biol Chem 286: 1567-1575
- PubMed: 21047786
- DOI: https://doi.org/10.1074/jbc.M110.144006
- Primary Citation of Related Structures:
3OZQ - PubMed Abstract:
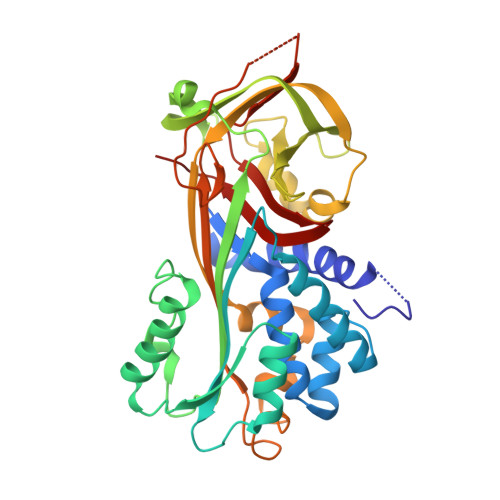
The Toll signaling pathway, an essential innate immune response in invertebrates, is mediated via the serine protease cascade. Once activated, the serine proteases are irreversibly inactivated by serine protease inhibitors (serpins). Recently, we identified three serpin-serine protease pairs that are directly involved in the regulation of Toll signaling cascade in a large beetle, Tenebrio molitor. Of these, the serpin SPN48 was cleaved by its target serine protease, Spätzle-processing enzyme, at a noncanonical P1 residue of the serpin's reactive center loop. To address this unique cleavage, we report the crystal structure of SPN48, revealing that SPN48 exhibits a native conformation of human antithrombin, where the reactive center loop is partially inserted into the center of the largest β-sheet of SPN48. The crystal structure also shows that SPN48 has a putative heparin-binding site that is distinct from those of the mammalian serpins. Ensuing biochemical studies demonstrate that heparin accelerates the inhibition of Spätzle-processing enzyme by a proximity effect in targeting the SPN48. Our finding provides the molecular mechanism of how serpins tightly regulate innate immune responses in invertebrates.
Organizational Affiliation:
College of Pharmacy and Research Institute for Drug Development, Pusan National University, Jangjeon-dong, Geumjeong-gu, Busan 609-735, Republic of Korea.