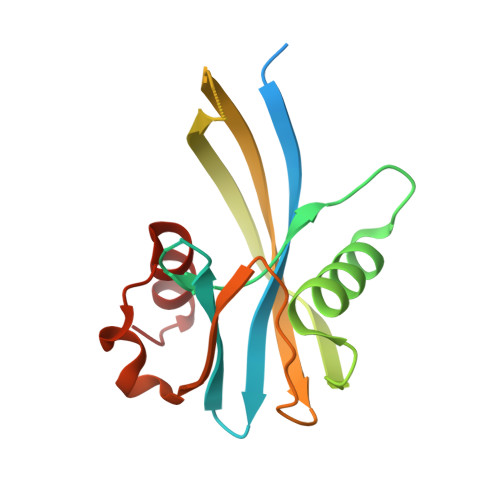
1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2
Halavaty, A.S., Minasov, G., Shuvalova, L., Winsor, J., Dubrovska, I., Peterson, S., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.