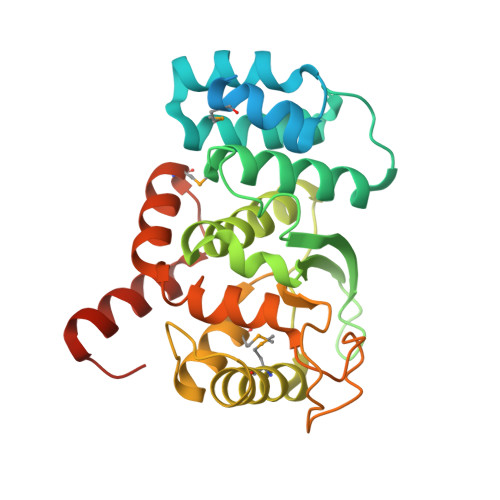
Novel structure of an N-terminal domain that is crucial for the dimeric assembly and DNA-binding of an archaeal DNA polymerase D large subunit from Pyrococcus horikoshii
Matsui, I., Urushibata, Y., Shen, Y., Matsui, E., Yokoyama, H.(2011) FEBS Lett 585: 452-458
- PubMed: 21192935
- DOI: https://doi.org/10.1016/j.febslet.2010.12.040
- Primary Citation of Related Structures:
3O59 - PubMed Abstract:
Archaea-specific D-family DNA polymerase forms a heterotetramer consisting of two large polymerase subunits and two small exonuclease subunits. The N-terminal (1-300) domain structure of the large subunit was determined by X-ray crystallography, although ∼50 N-terminal residues were disordered. The determined structure consists of nine alpha helices and three beta strands. We also identified the DNA-binding ability of the domain by SPR measurement. The N-terminal (1-100) region plays crucial roles in the folding of the large subunit dimer by connecting the ∼50 N-terminal residues with their own catalytic region (792-1163).
Organizational Affiliation:
Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan. ik-matsui@aist.go.jp