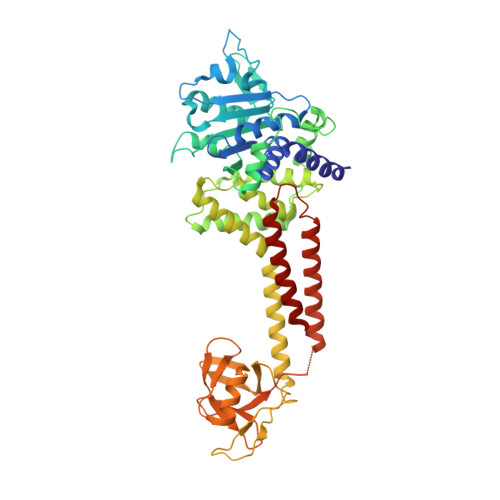
Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity
El Bakkouri, M., Gutsche, I., Kanjee, U., Zhao, B., Yu, M., Goret, G., Schoehn, G., Burmeister, W.P., Houry, W.A.(2010) Proc Natl Acad Sci U S A 107: 22499-22504
- PubMed: 21148420
- DOI: https://doi.org/10.1073/pnas.1009092107
- Primary Citation of Related Structures:
3NBX - PubMed Abstract:
The MoxR family of AAA+ ATPases is widespread throughout bacteria and archaea but remains poorly characterized. We recently found that the Escherichia coli MoxR protein, RavA (Regulatory ATPase variant A), tightly interacts with the inducible lysine decarboxylase, LdcI/CadA, to form a unique cage-like structure. Here, we present the X-ray structure of RavA and show that the αβα and all-α subdomains in the RavA AAA+ module are arranged as in magnesium chelatases rather than as in classical AAA+ proteins. RavA structure also contains a discontinuous triple-helical domain as well as a β-barrel-like domain forming a unique fold, which we termed the LARA domain. The LARA domain was found to mediate the interaction between RavA and LdcI. The RavA structure provides insights into how five RavA hexamers interact with two LdcI decamers to form the RavA-LdcI cage-like structure.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto, ON, Canada M5S 1A8.