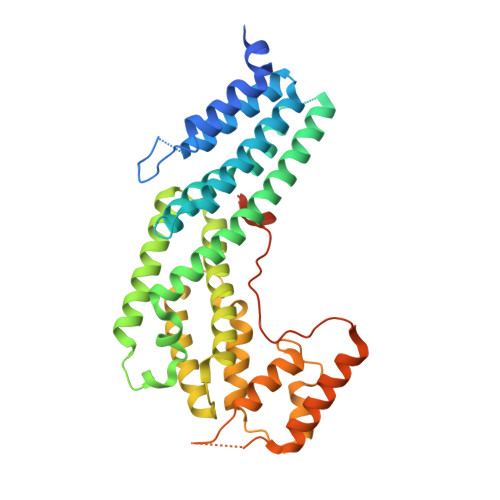
The structure of the Myo4p globular tail and its function in ASH1 mRNA localization.
Heuck, A., Fetka, I., Brewer, D.N., Huls, D., Munson, M., Jansen, R.P., Niessing, D.(2010) J Cell Biol 189: 497-510
- PubMed: 20439999
- DOI: https://doi.org/10.1083/jcb.201002076
- Primary Citation of Related Structures:
3MMI - PubMed Abstract:
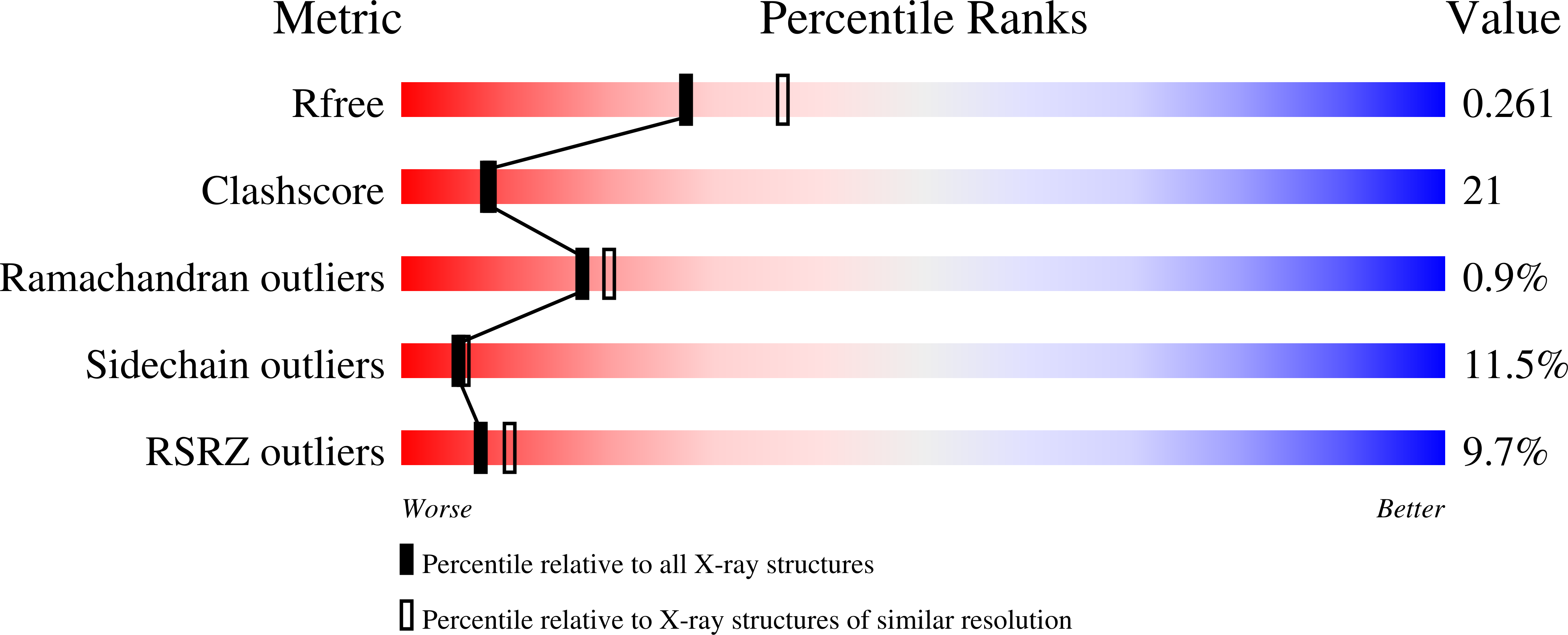
Type V myosin (MyoV)-dependent transport of cargo is an essential process in eukaryotes. Studies on yeast and vertebrate MyoV showed that their globular tails mediate binding to the cargo complexes. In Saccharomyces cerevisiae, the MyoV motor Myo4p interacts with She3p to localize asymmetric synthesis of HO 1 (ASH1) mRNA into the bud of dividing cells. A recent study showed that localization of GFP-MS2-tethered ASH1 particles does not require the Myo4p globular tail, challenging the supposed role of this domain. We assessed ASH1 mRNA and Myo4p distribution more directly and found that their localization is impaired in cells expressing globular tail-lacking Myo4p. In vitro studies further show that the globular tail together with a more N-terminal linker region is required for efficient She3p binding. We also determined the x-ray structure of the Myo4p globular tail and identify a conserved surface patch important for She3p binding. The structure shows pronounced similarities to membrane-tethering complexes and indicates that Myo4p may not undergo auto-inhibition of its motor domain.
Organizational Affiliation:
Institute of Structural Biology, Helmholtz Zentrum München, German Research Center for Environmental Health, Munich, Germany.