Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19
Schmidt, F., Treiber, N., Zocher, G., Bjelic, S., Steinmetz, M.O., Kalbacher, H., Stehle, T., Dodt, G.(2010) J Biol Chem 285: 25410-25417
- PubMed: 20554521
- DOI: https://doi.org/10.1074/jbc.M110.138503
- Primary Citation of Related Structures:
3MK4 - PubMed Abstract:
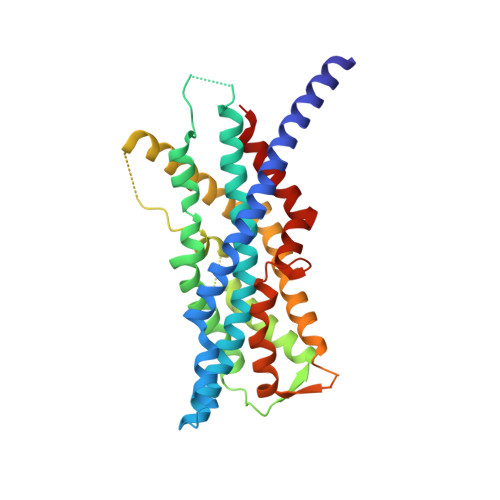
The human peroxins PEX3 and PEX19 play a central role in peroxisomal membrane biogenesis. The membrane-anchored PEX3 serves as the receptor for cytosolic PEX19, which in turn recognizes newly synthesized peroxisomal membrane proteins. After delivering these proteins to the peroxisomal membrane, PEX19 is recycled to the cytosol. The molecular mechanisms underlying these processes are not well understood. Here, we report the crystal structure of the cytosolic domain of PEX3 in complex with a PEX19-derived peptide. PEX3 adopts a novel fold that is best described as a large helical bundle. A hydrophobic groove at the membrane-distal end of PEX3 engages the PEX19 peptide with nanomolar affinity. Mutagenesis experiments identify phenylalanine 29 in PEX19 as critical for this interaction. Because key PEX3 residues involved in complex formation are highly conserved across species, the observed binding mechanism is of general biological relevance.
Organizational Affiliation:
Interfaculty Institute for Biochemistry, University of Tübingen, 72076 Tübingen, Germany.