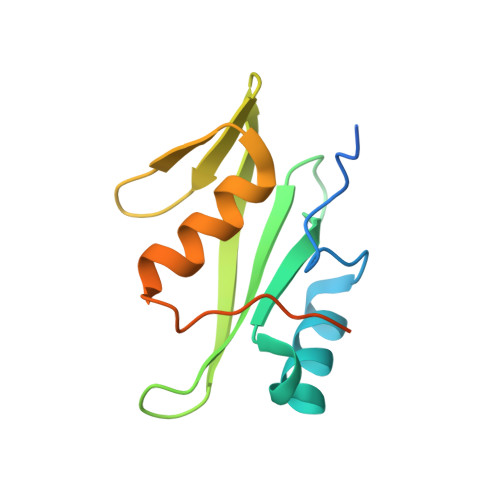
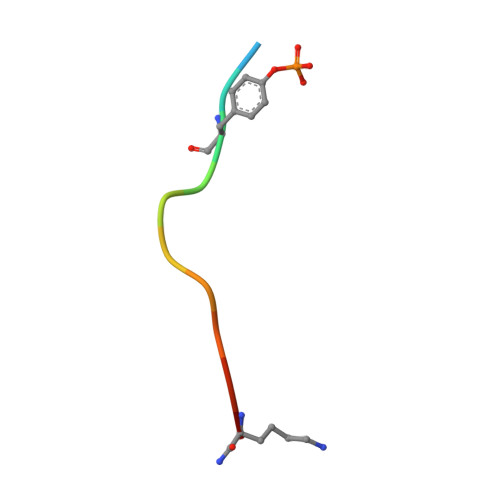
Loops govern SH2 domain specificity by controlling access to binding pockets.
Kaneko, T., Huang, H., Zhao, B., Li, L., Liu, H., Voss, C.K., Wu, C., Schiller, M.R., Li, S.S.(2010) Sci Signal 3: ra34-ra34
- PubMed: 20442417
- DOI: https://doi.org/10.1126/scisignal.2000796
- Primary Citation of Related Structures:
3MAZ - PubMed Abstract:
Cellular functions require specific protein-protein interactions that are often mediated by modular domains that use binding pockets to engage particular sequence motifs in their partners. Yet, how different members of a domain family select for distinct sequence motifs is not fully understood. The human genome encodes 120 Src homology 2 (SH2) domains (in 110 proteins), which mediate protein-protein interactions by binding to proteins with diverse phosphotyrosine (pTyr)-containing sequences. The structure of the SH2 domain of BRDG1 bound to a peptide revealed a binding pocket that was blocked by a loop residue in most other SH2 domains. Analysis of 63 SH2 domain structures suggested that the SH2 domains contain three binding pockets, which exhibit selectivity for the three positions after the pTyr in a peptide, and that SH2 domain loops defined the accessibility and shape of these pockets. Despite sequence variability in the loops, we identified conserved structural features in the loops of SH2 domains responsible for controlling access to these surface pockets. We engineered new loops in an SH2 domain that altered specificity as predicted. Thus, selective blockage of binding subsites or pockets by surface loops provides a molecular basis by which the diverse modes of ligand recognition by the SH2 domain may have evolved and provides a framework for engineering SH2 domains and designing SH2-specific inhibitors.
Organizational Affiliation:
Department of Biochemistry and the Siebens-Drake Research Institute, Schulich School of Medicine and Dentistry, University of Western Ontario, London, Ontario, Canada N6A 5C1.