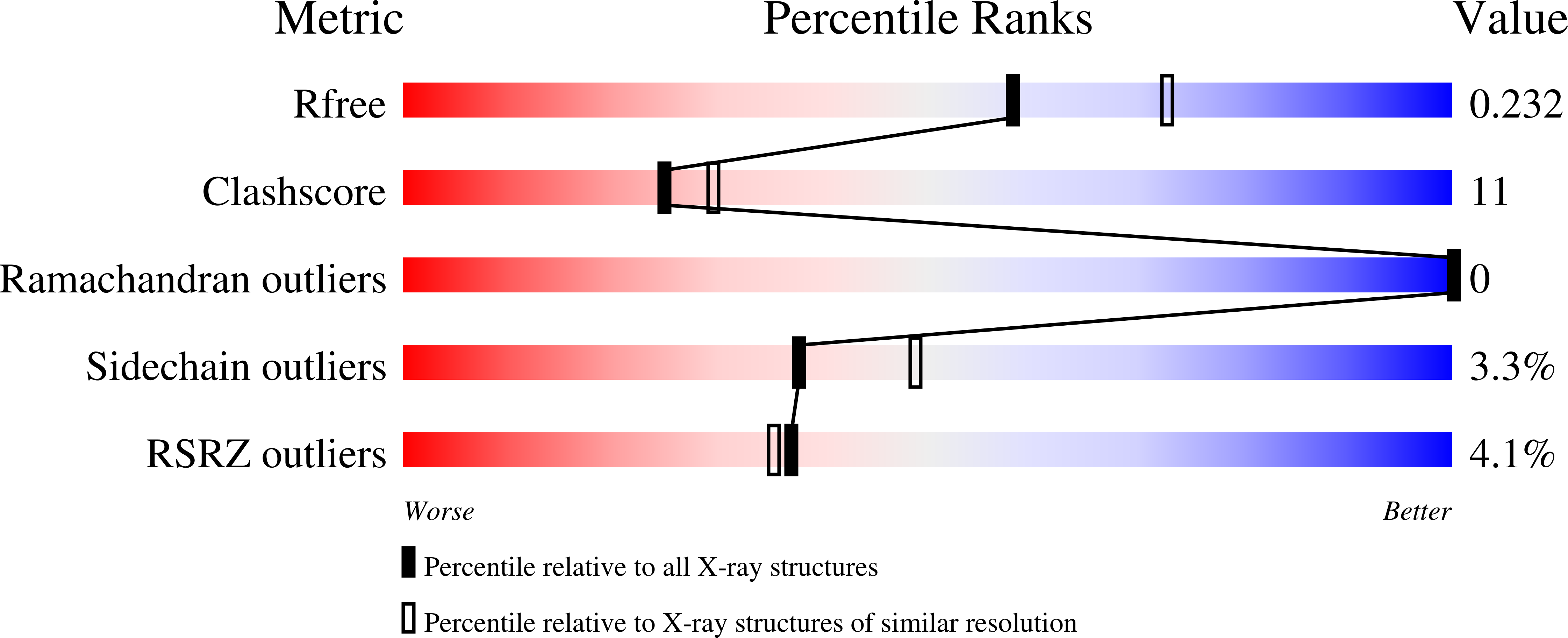
Structural and functional conservation of Mycobacterium tuberculosis GroEL paralogs suggests that GroEL1 Is a chaperonin.
Sielaff, B., Lee, K.S., Tsai, F.T.(2011) J Mol Biol 405: 831-839
- PubMed: 21094166
- DOI: https://doi.org/10.1016/j.jmb.2010.11.021
- Primary Citation of Related Structures:
3M6C - PubMed Abstract:
GroEL is a group I chaperonin that facilitates protein folding and prevents protein aggregation in the bacterial cytosol. Mycobacteria are unusual in encoding two or more copies of GroEL in their genome. While GroEL2 is essential for viability and likely functions as the general housekeeping chaperonin, GroEL1 is dispensable, but its structure and function remain unclear. Here, we present the 2.2-Å resolution crystal structure of a 23-kDa fragment of Mycobacterium tuberculosis GroEL1 consisting of an extended apical domain. Our X-ray structure of the GroEL1 apical domain closely resembles those of Escherichia coli GroEL and M. tuberculosis GroEL2, thus highlighting the remarkable structural conservation of bacterial chaperonins. Notably, in our structure, the proposed substrate-binding site of GroEL1 interacts with the N-terminal region of a symmetry-related neighboring GroEL1 molecule. The latter is consistent with the known GroEL apical domain function in substrate binding and is supported by results obtained from using peptide array technology. Taken together, these data show that the apical domains of M. tuberculosis GroEL paralogs are conserved in three-dimensional structure, suggesting that GroEL1, like GroEL2, is a chaperonin.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.