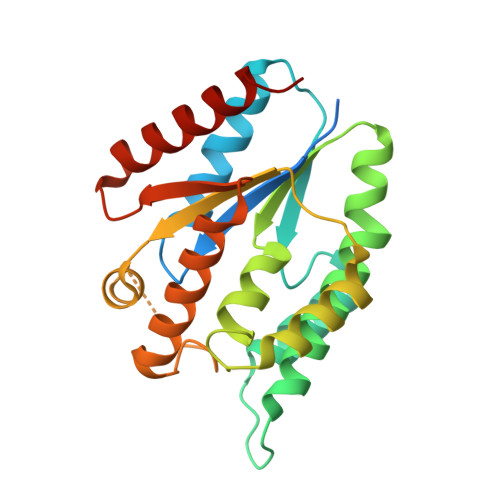
1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP
Halavaty, A.S., Minasov, G., Dubrovska, I., Winsor, J., Shuvalova, L., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.