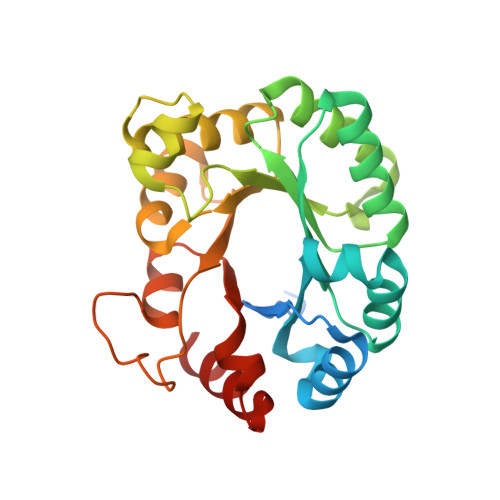
1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961
Halavaty, A.S., Shuvalova, L., Minasov, G., Dubrovska, I., Winsor, J., Glass, E.M., Peterson, S.N., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.