Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
Lu, W., Du, J., Stahl, M., Tzivelekidis, T., Belyi, Y., Gerhardt, S., Aktories, K., Einsle, O.(2009) J Mol Biol
- PubMed: 19941871
- DOI: https://doi.org/10.1016/j.jmb.2009.11.044
- Primary Citation of Related Structures:
3JSZ, 3JT1 - PubMed Abstract:
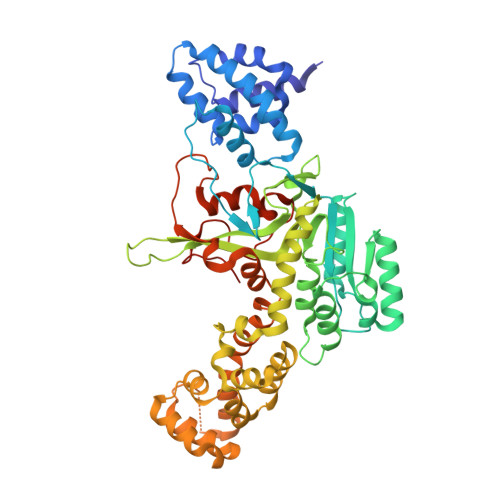
The glucosyltransferase Lgt1 is one of three glucosylating toxins of Legionella pneumophila, the causative agent of Legionnaires disease. It acts through specific glucosylation of a serine residue (S53) in the eukaryotic elongation factor 1A and belongs to type A glycosyltransferases. High-resolution crystal structures of Lgt1 show an elongated shape of the protein, with the binding site for uridine disphosphate glucose at the bottom of a deep cleft. Lgt1 shows only a low sequence identity with other type A glycosyltransferases, and structural conservation is limited to a central folding core that is usually observed within this family of proteins. Domains and protrusions added to the core motif represent determinants for the specific recognition and binding of the target. Manual docking experiments based on the crystal structures of toxin and target protein suggest an obvious mode of binding to the target that allows for efficient transfer of a glucose moiety.
Organizational Affiliation:
Institut für organische Chemie und Biochemie, Albert-Ludwigs-Universität Freiburg, D-79104 Freiburg, Germany.