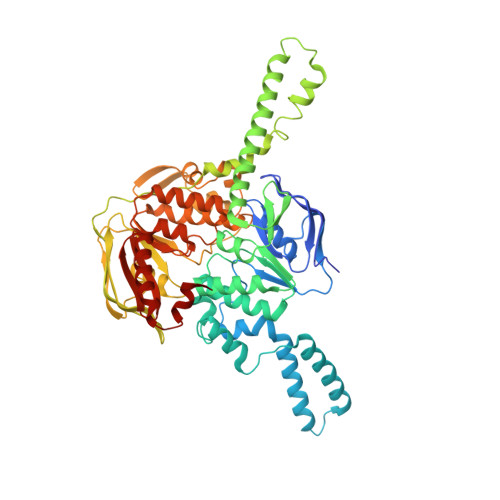
EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics.
Chen, B., Boel, G., Hashem, Y., Ning, W., Fei, J., Wang, C., Gonzalez, R.L., Hunt, J.F., Frank, J.(2014) Nat Struct Mol Biol 21: 152-159
- PubMed: 24389465
- DOI: https://doi.org/10.1038/nsmb.2741
- Primary Citation of Related Structures:
3J5S - PubMed Abstract:
Cells express many ribosome-interacting factors whose functions and molecular mechanisms remain unknown. Here, we elucidate the mechanism of a newly characterized regulatory translation factor, energy-dependent translational throttle A (EttA), which is an Escherichia coli representative of the ATP-binding cassette F (ABC-F) protein family. Using cryo-EM, we demonstrate that the ATP-bound form of EttA binds to the ribosomal tRNA-exit site, where it forms bridging interactions between the ribosomal L1 stalk and the tRNA bound in the peptidyl-tRNA-binding site. Using single-molecule fluorescence resonance energy transfer, we show that the ATP-bound form of EttA restricts ribosome and tRNA dynamics required for protein synthesis. This work represents the first example, to our knowledge, in which the detailed molecular mechanism of any ABC-F family protein has been determined and establishes a framework for elucidating the mechanisms of other regulatory translation factors.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York, USA.