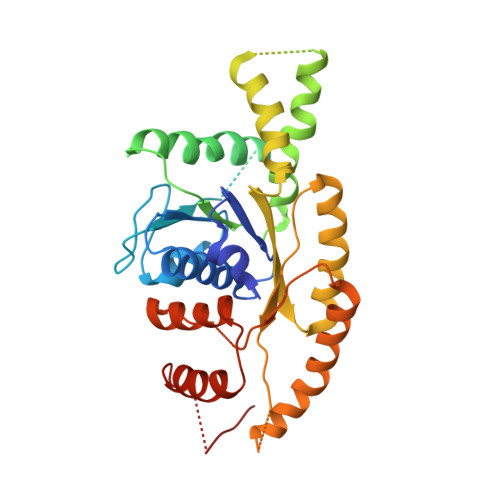
Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Suloway, C.J., Chartron, J.W., Zaslaver, M., Clemons, W.M.(2009) Proc Natl Acad Sci U S A 106: 14849-14854
- PubMed: 19706470
- DOI: https://doi.org/10.1073/pnas.0907522106
- Primary Citation of Related Structures:
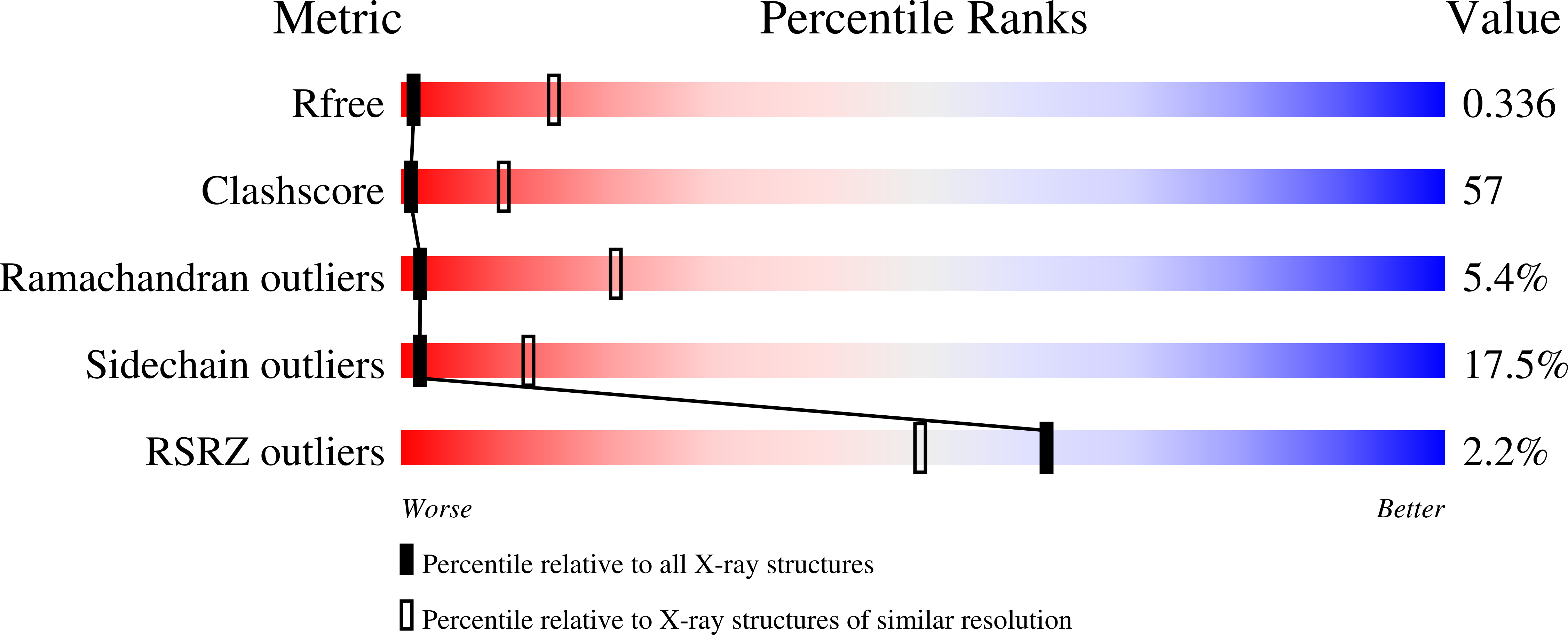
3IBG, 3IDQ - PubMed Abstract:
The Get3 ATPase directs the delivery of tail-anchored (TA) proteins to the endoplasmic reticulum (ER). TA-proteins are characterized by having a single transmembrane helix (TM) at their extreme C terminus and include many essential proteins, such as SNAREs, apoptosis factors, and protein translocation components. These proteins cannot follow the SRP-dependent co-translational pathway that typifies most integral membrane proteins; instead, post-translationally, these proteins are recognized and bound by Get3 then delivered to the ER in the ATP dependent Get pathway. To elucidate a molecular mechanism for TA protein binding by Get3 we have determined three crystal structures in apo and ADP forms from Saccharomyces cerevisae (ScGet3-apo) and Aspergillus fumigatus (AfGet3-apo and AfGet3-ADP). Using structural information, we generated mutants to confirm important interfaces and essential residues. These results point to a model of how Get3 couples ATP hydrolysis to the binding and release of TA-proteins.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, CA 91125, USA.