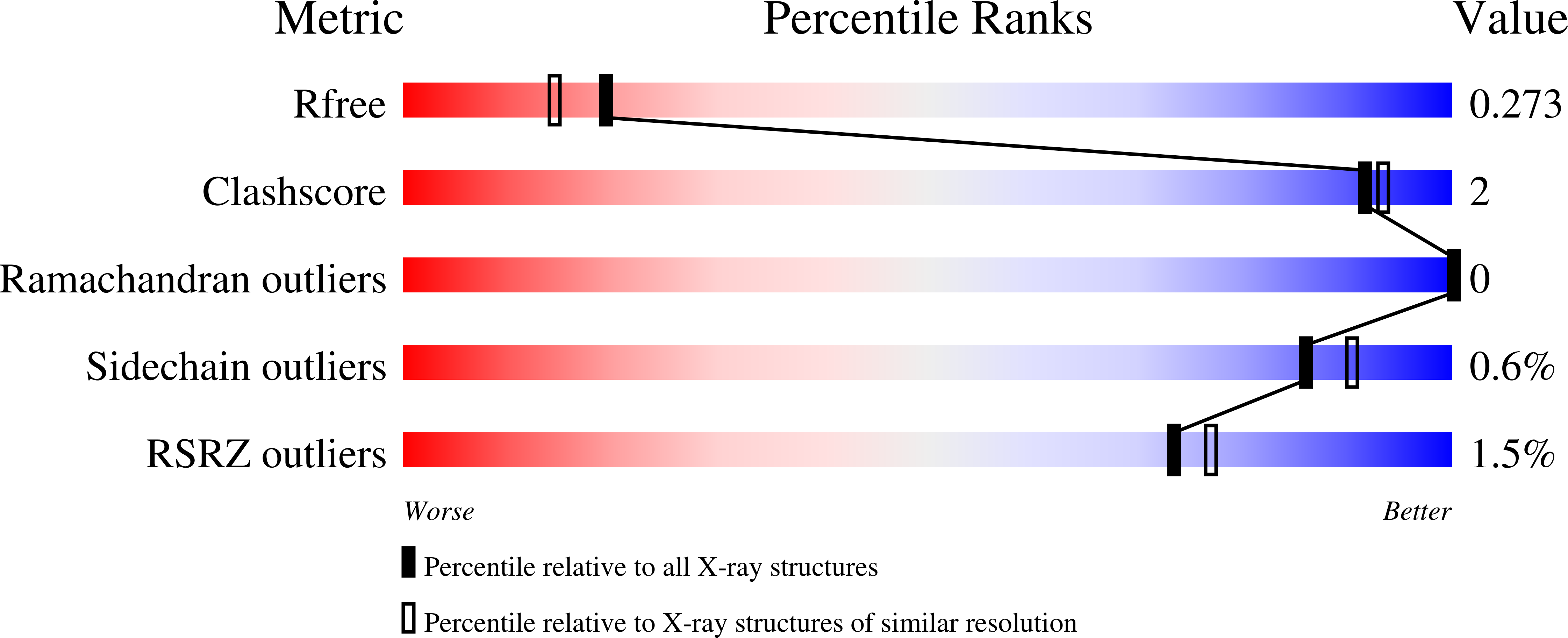
Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
Liu, W., Zhao, R., McFarland, C., Kieft, J., Niedzwiecka, A., Jankowska-Anyszka, M., Stepinski, J., Darzynkiewicz, E., Jones, D.N., Davis, R.E.(2009) J Biol Chem 284: 31336-31349
- PubMed: 19710013
- DOI: https://doi.org/10.1074/jbc.M109.049858
- Primary Citation of Related Structures:
3HXG, 3HXI - PubMed Abstract:
The eukaryotic translation initiation factor eIF4E recognizes the mRNA cap, a key step in translation initiation. Here we have characterized eIF4E from the human parasite Schistosoma mansoni. Schistosome mRNAs have either the typical monomethylguanosine (m(7)G) or a trimethylguanosine (m(2,2,7)G) cap derived from spliced leader trans-splicing. Quantitative fluorescence titration analyses demonstrated that schistosome eIF4E has similar binding specificity for both caps. We present the first crystal structure of an eIF4E with similar binding specificity for m(7)G and m(2,2,7)G caps. The eIF4E.m(7)GpppG structure demonstrates that the schistosome protein binds monomethyl cap in a manner similar to that of single specificity eIF4Es and exhibits a structure similar to other known eIF4Es. The structure suggests an alternate orientation of a conserved, key Glu-90 in the cap-binding pocket that may contribute to dual binding specificity and a position for mRNA bound to eIF4E consistent with biochemical data. Comparison of NMR chemical shift perturbations in schistosome eIF4E on binding m(7)GpppG and m(2,2,7)GpppG identified key differences between the two complexes. Isothermal titration calorimetry demonstrated significant thermodynamics differences for the binding process with the two caps (m(7)G versus m(2,2,7)G). Overall the NMR and isothermal titration calorimetry data suggest the importance of intrinsic conformational flexibility in the schistosome eIF4E that enables binding to m(2,2,7)G cap.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Colorado School of Medicine, Aurora, Colorado 80045, USA.