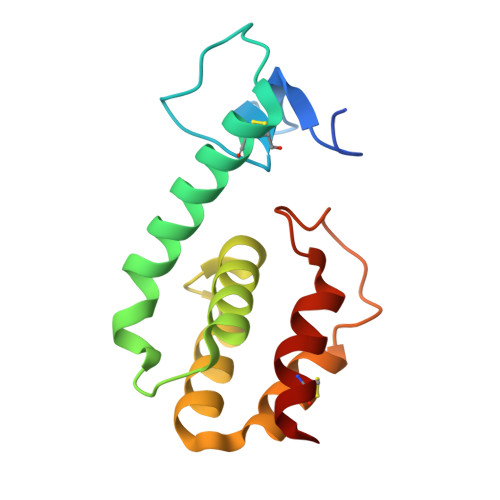
Regulation of a muralytic enzyme by dynamic membrane topology.
Sun, Q., Kuty, G.F., Arockiasamy, A., Xu, M., Young, R., Sacchettini, J.C.(2009) Nat Struct Mol Biol 16: 1192-1194
- PubMed: 19881499
- DOI: https://doi.org/10.1038/nsmb.1681
- Primary Citation of Related Structures:
3HDE, 3HDF - PubMed Abstract:
R(21), the lysozyme of coliphage 21, has an N-terminal signal-anchor-release (SAR) domain that directs its secretion in a membrane-tethered, inactive form and then its release and activation in the periplasm. Both genetic and crystallographic studies show that the SAR domain, once extracted from the bilayer, refolds into the body of the enzyme and effects muralytic activation by repositioning one residue of the canonical lysozyme catalytic triad.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas, USA.