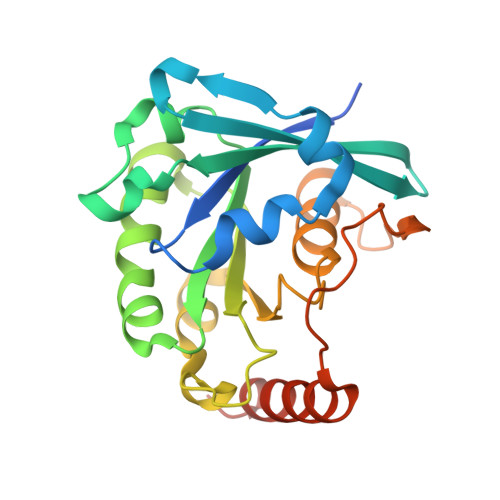
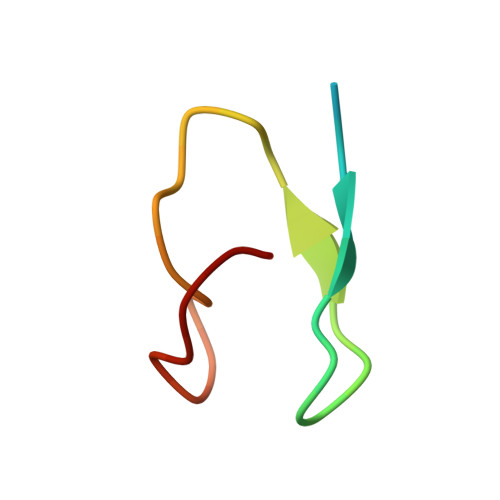
Crystallographic and Biochemical Analysis of the Ran-binding Zinc Finger Domain.
Partridge, J.R., Schwartz, T.U.(2009) J Mol Biol 391: 375-389
- PubMed: 19505478
- DOI: https://doi.org/10.1016/j.jmb.2009.06.011
- Primary Citation of Related Structures:
3GJ0, 3GJ3, 3GJ4, 3GJ5, 3GJ6, 3GJ7, 3GJ8 - PubMed Abstract:
The nuclear pore complex (NPC) resides in circular openings within the nuclear envelope and serves as the sole conduit to facilitate nucleocytoplasmic transport in eukaryotes. The asymmetric distribution of the small G protein Ran across the nuclear envelope regulates directionality of protein transport. Ran interacts with the NPC of metazoa via two asymmetrically localized components, Nup153 at the nuclear face and Nup358 at the cytoplasmic face. Both nucleoporins contain a stretch of distinct, Ran-binding zinc finger domains. Here, we present six crystal structures of Nup153-zinc fingers in complex with Ran and a 1.48 A crystal structure of RanGDP. Crystal engineering allowed us to obtain well diffracting crystals so that all ZnF-Ran complex structures are refined to high resolution. Each of the four zinc finger modules of Nup153 binds one Ran molecule in apparently non-allosteric fashion. The affinity is measurably higher for RanGDP than for RanGTP and varies modestly between the individual zinc fingers. By microcalorimetric and mutational analysis, we determined that one specific hydrogen bond accounts for most of the differences in the binding affinity of individual zinc fingers. Genomic analysis reveals that only in animals do NPCs contain Ran-binding zinc fingers. We speculate that these organisms evolved a mechanism to maintain a high local concentration of Ran at the vicinity of the NPC, using this zinc finger domain as a sink.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.