Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1
Malashkevich, V.N., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase | 478 | Methanosarcina mazei Go1 | Mutation(s): 0 Gene Names: MM_1154 EC: 1.1.1 | 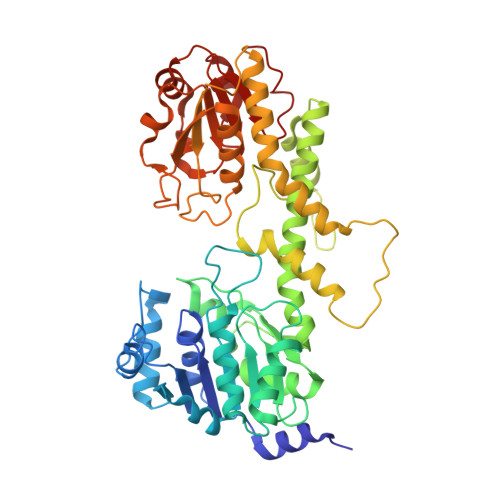 | |
UniProt | |||||
Find proteins for Q8PXR2 (Methanosarcina mazei (strain ATCC BAA-159 / DSM 3647 / Goe1 / Go1 / JCM 11833 / OCM 88)) Explore Q8PXR2 Go to UniProtKB: Q8PXR2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8PXR2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.83 | α = 90 |
| b = 88.723 | β = 90 |
| c = 151.6 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| RESOLVE | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| CBASS | data collection |
| HKL-2000 | data reduction |
| PHENIX | phasing |