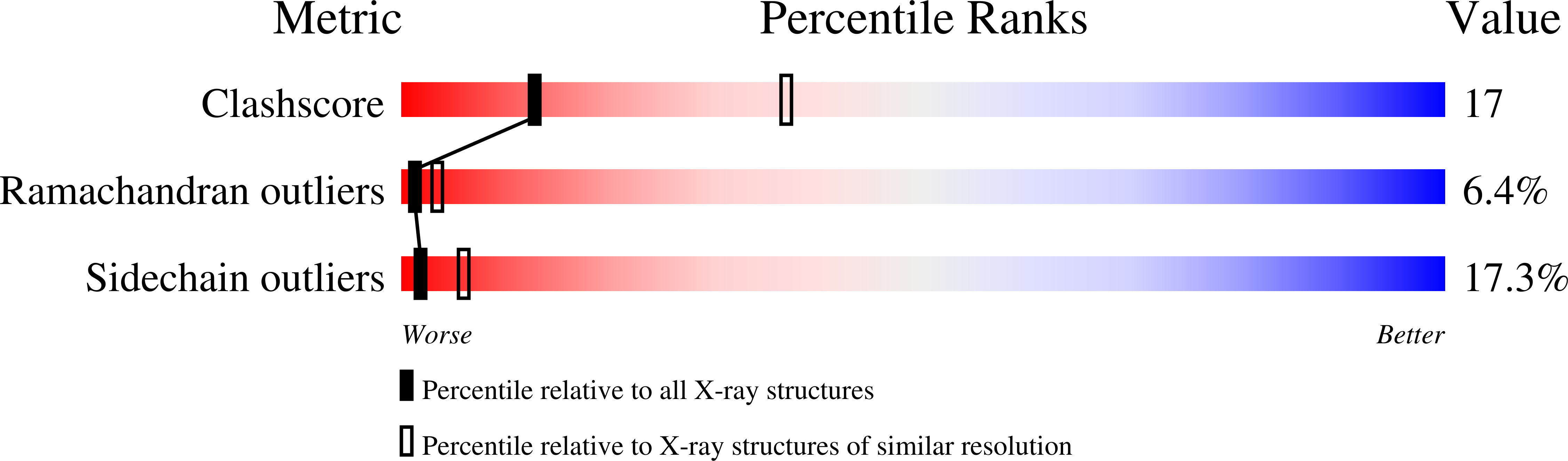Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
Ke, H.M., Thorpe, C.M., Seaton, B., Lipscomb, W.N., Marcus, F.(1990) J Mol Biol 212: 513-539
- PubMed: 2157849
- DOI: https://doi.org/10.1016/0022-2836(90)90329-k
- Primary Citation of Related Structures:
2FBP, 3FBP - PubMed Abstract:
The structures of the native fructose-1,6-bisphosphatase (Fru-1,6-Pase), from pig kidney cortex, and its fructose 2,6-bisphosphate (Fru-2,6-P2) complexes have been refined to 2.8 A resolution to R-factors of 0.194 and 0.188, respectively. The root-mean-square deviations from the standard geometry are 0.021 A and 0.016 A for the bond length, and 4.4 degrees and 3.8 degrees for the bond angle. Four sites for Fru-2,6-P2 binding per tetramer have been identified by difference Fourier techniques. The Fru-2,6-P2 site has the shape of an oval cave about 10 A deep, and with other dimensions about 18 A by 12 A. The two Fru-2,6-P2 binding caves of the dimer in the crystallographically asymmetric unit sit next to one another and open in opposite directions. These two binding sites mutually exchange their Arg243 side-chains, indicating the potential for communication between the two sites. The beta, D-fructose 2,6-bisphosphate has been built into the density and refined well. The oxygen atoms of the 6-phosphate group of Fru-2,6-P2 interact with Arg243 from the adjacent monomer and the residues of Lys274, Asn212, Tyr264, Tyr215 and Tyr244 in the same monomer. The sugar ring primarily contacts with the backbone atoms from Gly246 to Met248, as well as the side-chain atoms, Asp121, Glu280 and Lys274. The 2-phosphate group interacts with the side-chain atoms of Ser124 and Lys274. A negatively charged pocket near the 2-phosphate group includes Asp118, Asp121 and Glu280, as well as Glu97 and Glu98. The 2-phosphate group showed a disordered binding perhaps because of the disturbance from the negatively charged pocket. In addition, Asn125 and Lys269 are located within a 5 A radius of Fru-2,6-P2. We argue that Fru-2,6-P2 binds to the active site of the enzyme on the basis of the following observations: (1) the structure similarity between Fru-2,6-P2 and the substrate; (2) sequence conservation of the residues directly interacting with Fru-2,6-P2 or located at the negatively charged pocket; (3) a divalent metal site next to the 2-phosphate group of Fru-2,6-P2; and (4) identification of some active site residues in our structure, e.g. tyrosine and Lys274, consistent with the results of the ultraviolet spectra and the chemical modification. The structures are described in detail including interactions of interchain surfaces, and the chemically modifiable residues are discussed on the basis of the refined structures.(ABSTRACT TRUNCATED AT 400 WORDS)
Organizational Affiliation:
Gibbs Chemical Laboratory, Harvard University, Cambridge, MA 02138.