Probing the function of heme distortion in the H-NOX family.
Olea, C., Boon, E.M., Pellicena, P., Kuriyan, J., Marletta, M.A.(2008) ACS Chem Biol 3: 703-710
- PubMed: 19032091
- DOI: https://doi.org/10.1021/cb800185h
- Primary Citation of Related Structures:
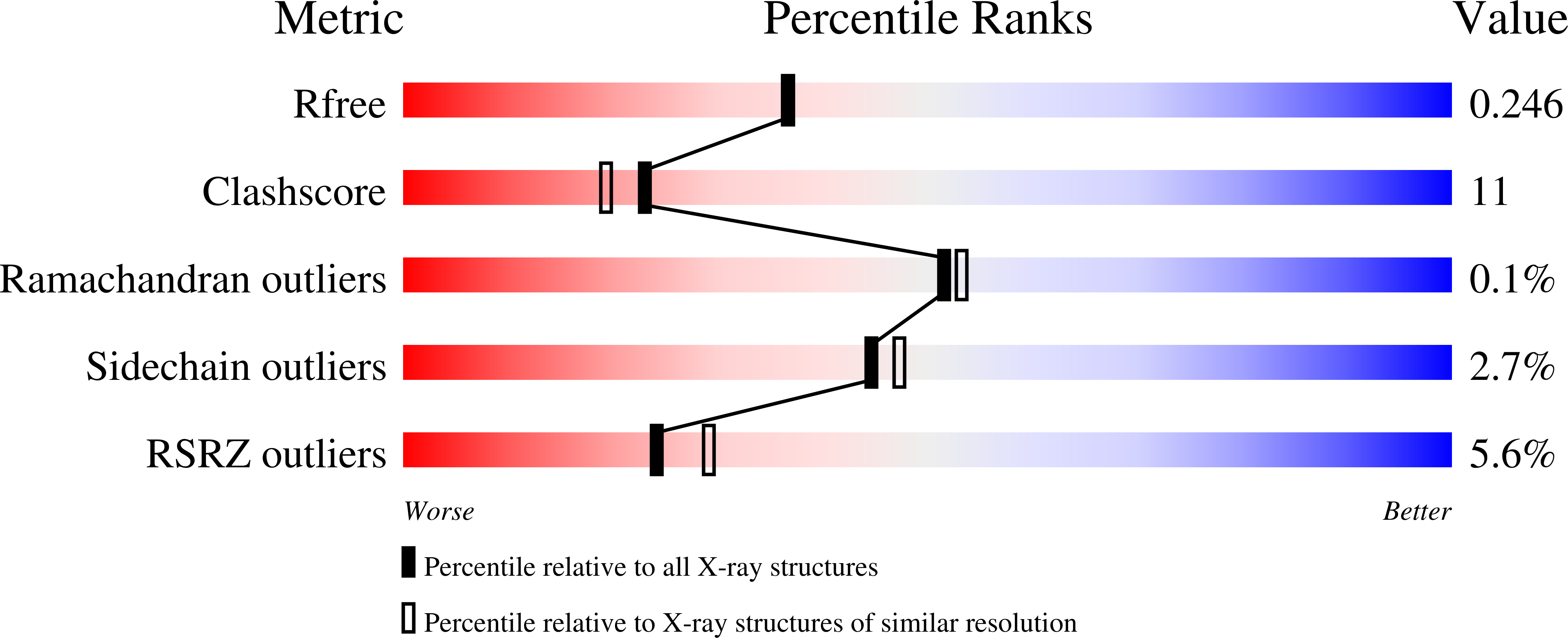
3EEE - PubMed Abstract:
Hemoproteins carry out diverse functions utilizing a wide range of chemical reactivity while employing the same heme prosthetic group. It is clear from high-resolution crystal structures and biochemical studies that protein-bound hemes are not planar and adopt diverse conformations. The crystal structure of an H-NOX domain from Thermoanaerobacter tengcongensis (Tt H-NOX) contains the most distorted heme reported to date. In this study, Tt H-NOX was engineered to adopt a flatter heme by mutating proline 115, a conserved residue in the H-NOX family, to alanine. Decreasing heme distortion in Tt H-NOX increases affinity for oxygen and decreases the reduction potential of the heme iron. Additionally, flattening the heme is associated with significant shifts in the N-terminus of the protein. These results show a clear link between the heme conformation and Tt H-NOX structure and demonstrate that heme distortion is an important determinant for maintaining biochemical properties in H-NOX proteins.
Organizational Affiliation:
Department of Molecular and Cell Biology, California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA.