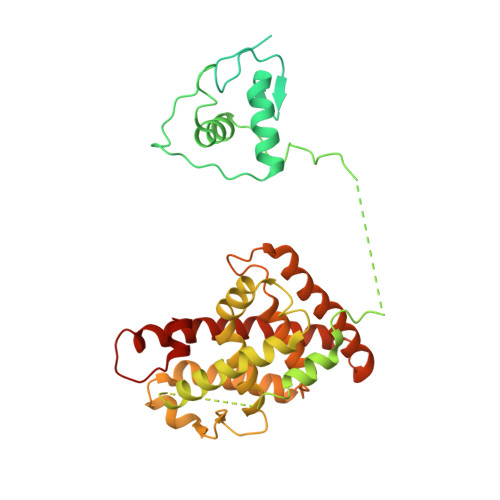
Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Chandra, V., Huang, P., Hamuro, Y., Raghuram, S., Wang, Y., Burris, T.P., Rastinejad, F.(2008) Nature 456: 350-356
- PubMed: 19043829
- DOI: https://doi.org/10.1038/nature07413
- Primary Citation of Related Structures:
3DZU, 3DZY, 3E00 - PubMed Abstract:
Nuclear receptors are multi-domain transcription factors that bind to DNA elements from which they regulate gene expression. The peroxisome proliferator-activated receptors (PPARs) form heterodimers with the retinoid X receptor (RXR), and PPAR-gamma has been intensively studied as a drug target because of its link to insulin sensitization. Previous structural studies have focused on isolated DNA or ligand-binding segments, with no demonstration of how multiple domains cooperate to modulate receptor properties. Here we present structures of intact PPAR-gamma and RXR-alpha as a heterodimer bound to DNA, ligands and coactivator peptides. PPAR-gamma and RXR-alpha form a non-symmetric complex, allowing the ligand-binding domain (LBD) of PPAR-gamma to contact multiple domains in both proteins. Three interfaces link PPAR-gamma and RXR-alpha, including some that are DNA dependent. The PPAR-gamma LBD cooperates with both DNA-binding domains (DBDs) to enhance response-element binding. The A/B segments are highly dynamic, lacking folded substructures despite their gene-activation properties.
Organizational Affiliation:
Department of Pharmacology, and Center for Molecular Design, University of Virginia Health System, 1300 Jefferson Park Avenue, Charlottesville, Virginia 22908-0735, USA.