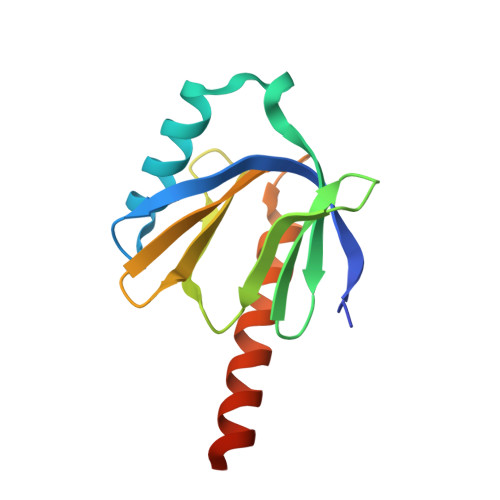
Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2.
Radzimanowski, J., Simon, B., Sattler, M., Beyreuther, K., Sinning, I., Wild, K.(2008) EMBO Rep 9: 1134-1140
- PubMed: 18833287
- DOI: https://doi.org/10.1038/embor.2008.188
- Primary Citation of Related Structures:
3DXC, 3DXD, 3DXE - PubMed Abstract:
Cleavage of the amyloid precursor protein (APP) is a crucial event in Alzheimer disease pathogenesis that creates the amyloid-beta peptide (Abeta) and liberates the carboxy-terminal APP intracellular domain (AICD) into the cytosol. The interaction of the APP C terminus with the adaptor protein Fe65 mediates APP trafficking and signalling, and is thought to regulate APP processing and Abeta generation. We determined the crystal structure of the AICD in complex with the C-terminal phosphotyrosine-binding (PTB) domain of Fe65. The unique interface involves the NPxY PTB-binding motif and two alpha helices. The amino-terminal helix of the AICD is capped by threonine T(668), an Alzheimer disease-relevant phosphorylation site involved in Fe65-binding regulation. The structure together with mutational studies, isothermal titration calorimetry and nuclear magnetic resonance experiments sets the stage for understanding T(668) phosphorylation-dependent complex regulation at a molecular level. A molecular switch model is proposed.
Organizational Affiliation:
Heidelberg University Biochemistry Center, University of Heidelberg, INF328, D-69120 Heidelberg, Germany.