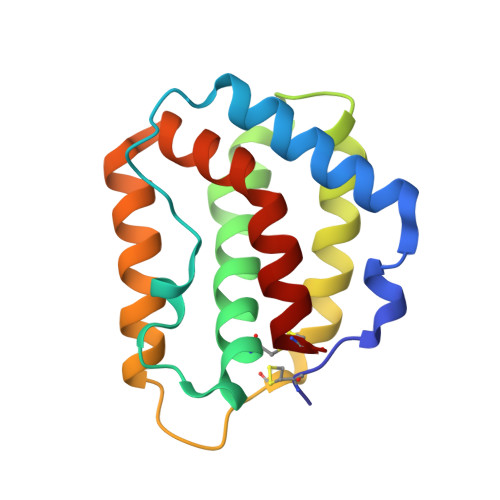
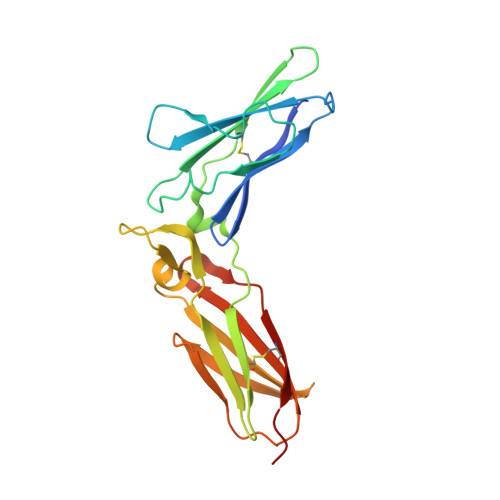
Structure of IL-22 Bound to Its High-Affinity IL-22R1 Chain.
Jones, B.C., Logsdon, N.J., Walter, M.R.(2008) Structure 16: 1333-1344
- PubMed: 18599299
- DOI: https://doi.org/10.1016/j.str.2008.06.005
- Primary Citation of Related Structures:
3DGC - PubMed Abstract:
IL-22 is an IL-10 family cytokine that initiates innate immune responses against bacterial pathogens and contributes to immune disease. IL-22 biological activity is initiated by binding to a cell-surface complex composed of IL-22R1 and IL-10R2 receptor chains and further regulated by interactions with a soluble binding protein, IL-22BP, which shares sequence similarity with an extracellular region of IL-22R1 (sIL-22R1). IL-22R1 also pairs with the IL-20R2 chain to induce IL-20 and IL-24 signaling. To define the molecular basis of these diverse interactions, we have determined the structure of the IL-22/sIL-22R1 complex. The structure, combined with homology modeling and surface plasmon resonance studies, defines the molecular basis for the distinct affinities and specificities of IL-22 and IL-10 receptor chains that regulate cellular targeting and signal transduction to elicit effective immune responses.
Organizational Affiliation:
Center for Biophysical Sciences and Engineering, University of Alabama at Birmingham, Birmingham, AL 35294, USA.