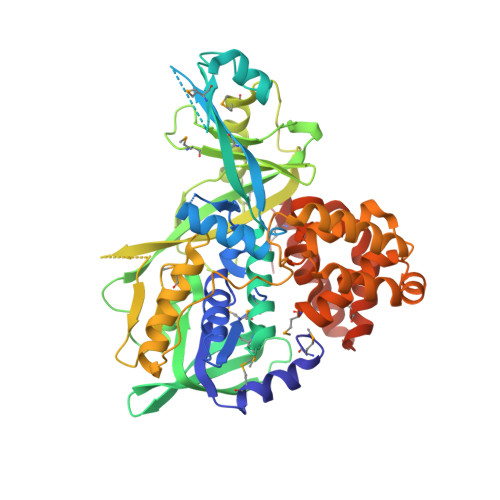
X-Ray structure of the glycerol-3-phosphate dehydrogenase from Bacillus halodurans complexed with FAD. Northeast Structural Genomics Consortium target BhR167.
Kuzin, A.P., Abashidze, M., Seetharaman, J., Wang, D., Janjua, H., Owens, L., Xiao, R., Nair, R., Baran, M.C., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.