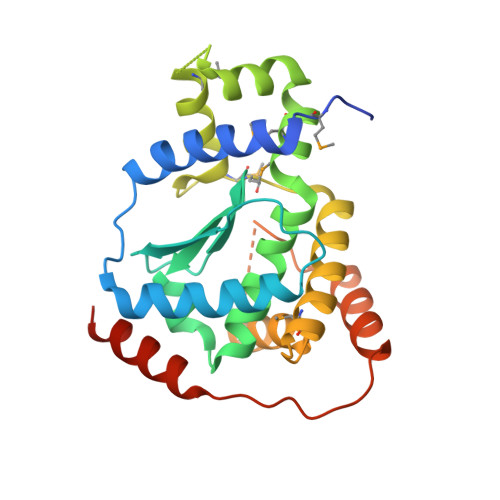
X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
Kuzin, A.P., Chen, Y., Seetharaman, J., Vorobiev, S.M., Forouhar, F., Wang, D., Mao, L., Maglaqui, M., Xiao, R., Liu, J., Baran, M.C., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.