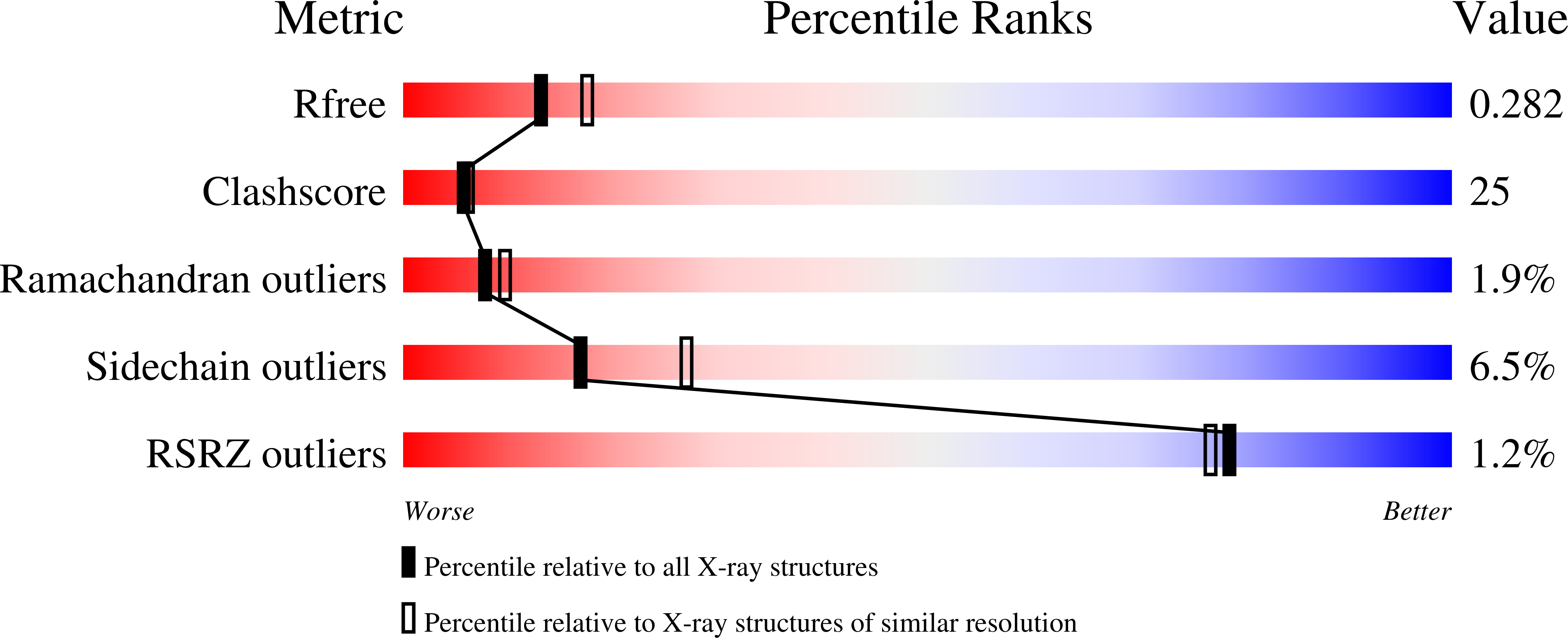
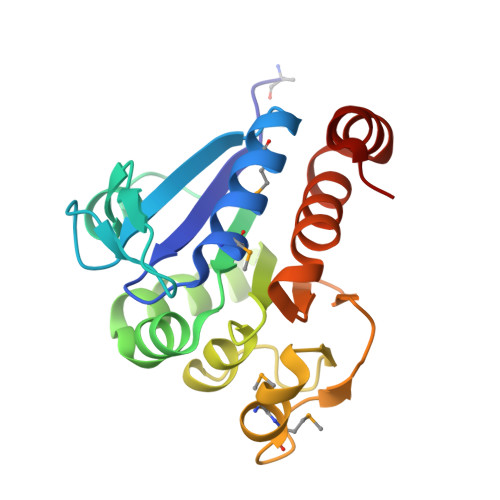
Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
Cha, S.S., Jung, H.I., Jeon, H., An, Y.J., Kim, I.K., Yun, S., Ahn, H.J., Chung, K.C., Lee, S.H., Suh, P.G., Kang, S.O.(2008) J Biol Chem 283: 34069-34075
- PubMed: 18922803
- DOI: https://doi.org/10.1074/jbc.M804243200
- Primary Citation of Related Structures:
3BWE - PubMed Abstract:
Mutations in the DJ-1 gene have been implicated in the autosomal recessive early onset parkinsonism. DJ-1 is a soluble dimeric protein with critical roles in response to oxidative stress and in neuronal maintenance. However, several lines of evidence suggest the existence of a nonfunctional aggregated form of DJ-1 in the brain of patients with some neurodegenerative diseases. Here, we show that inorganic phosphate, an important anion that exhibits elevated levels in patients with Parkinson disease, transforms DJ-1 into filamentous aggregates. According to the 2.4-A crystal structure, DJ-1 dimers are linearly stacked through P(i)-mediated interactions to form protofilaments, which are then bundled into a filamentous assembly.
Organizational Affiliation:
Marine and Extreme Genome Research Center, Korea Ocean Research & Development Institute, Ansan 426-744, Republic of Korea. chajung@kordi.re.kr