Crystal structure of the central axis DF complex of the prokaryotic V-ATPase
Saijo, S., Arai, S., Hossain, K.M.M., Yamato, I., Suzuki, K., Kakinuma, Y., Ishizuka-Katsura, Y., Ohsawa, N., Terada, T., Shirouzu, M., Yokoyama, S., Iwata, S., Murata, T.(2011) Proc Natl Acad Sci U S A 108: 19955-19960
- PubMed: 22114184
- DOI: https://doi.org/10.1073/pnas.1108810108
- Primary Citation of Related Structures:
3AON - PubMed Abstract:
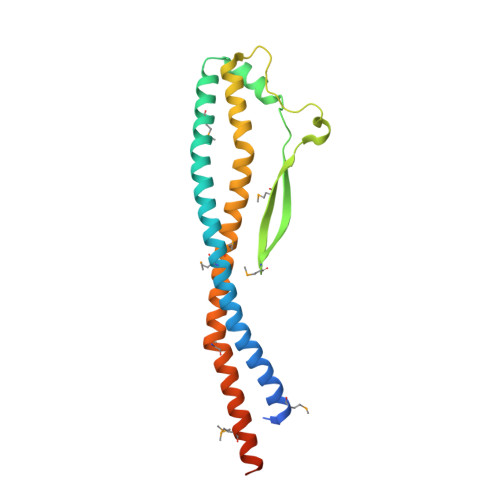
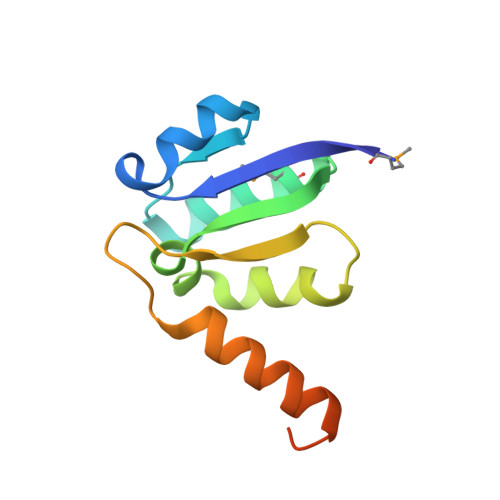
V-ATPases function as ATP-dependent ion pumps in various membrane systems of living organisms. ATP hydrolysis causes rotation of the central rotor complex, which is composed of the central axis D subunit and a membrane c ring that are connected by F and d subunits. Here we determined the crystal structure of the DF complex of the prokaryotic V-ATPase of Enterococcus hirae at 2.0-Å resolution. The structure of the D subunit comprised a long left-handed coiled coil with a unique short β-hairpin region that is effective in stimulating the ATPase activity of V(1)-ATPase by twofold. The F subunit is bound to the middle portion of the D subunit. The C-terminal helix of the F subunit, which was believed to function as a regulatory region by extending into the catalytic A(3)B(3) complex, contributes to tight binding to the D subunit by forming a three-helix bundle. Both D and F subunits are necessary to bind the d subunit that links to the c ring. From these findings, we modeled the entire rotor complex (DFdc ring) of V-ATPase.
Organizational Affiliation:
Department of Biological Science and Technology, Tokyo University of Science, 2641 Yamazaki, Noda-shi, Chiba 278-8510, Japan.