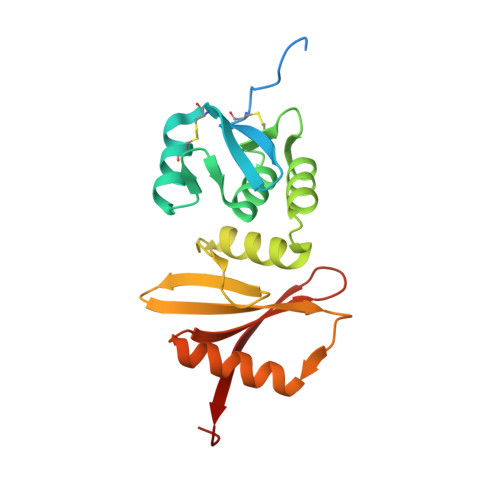
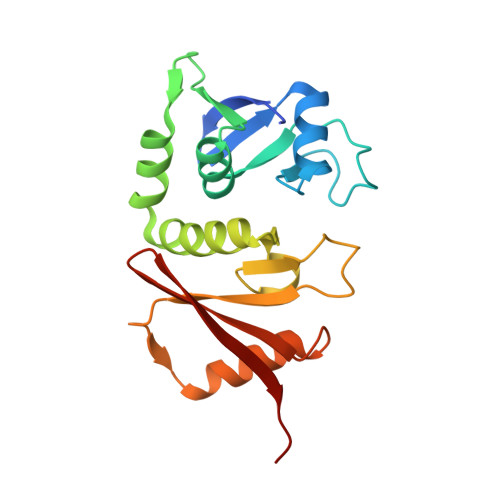
A Conserved Lysine Residue in the Crenarchaea-Specific Loop is Important for the Crenarchaeal Splicing Endonuclease Activity.
Okuda, M., Shiba, T., Inaoka, D.K., Kita, K., Kurisu, G., Mineki, S., Harada, S., Watanabe, Y., Yoshinari, S.(2011) J Mol Biol 405: 92-104
- PubMed: 21050862
- DOI: https://doi.org/10.1016/j.jmb.2010.10.050
- Primary Citation of Related Structures:
3AJV - PubMed Abstract:
In Archaea, splicing endonuclease (EndA) recognizes and cleaves precursor RNAs to remove introns. Currently, EndAs are classified into three families according to their subunit structures: homotetramer, homodimer, and heterotetramer. The crenarchaeal heterotetrameric EndAs can be further classified into two subfamilies based on the size of the structural subunit. Subfamily A possesses a structural subunit similar in size to the catalytic subunit, whereas subfamily B possesses a structural subunit significantly smaller than the catalytic subunit. Previously, we solved the crystal structure of an EndA from Pyrobaculum aerophilum. The endonuclease was classified into subfamily B, and the structure revealed that the enzyme lacks an N-terminal subdomain in the structural subunit. However, no structural information is available for crenarchaeal heterotetrameric EndAs that are predicted to belong to subfamily A. Here, we report the crystal structure of the EndA from Aeropyrum pernix, which is predicted to belong to subfamily A. The enzyme possesses the N-terminal subdomain in the structural subunit, revealing that the two subfamilies of heterotetrameric EndAs are structurally distinct. EndA from A. pernix also possesses an extra loop region that is characteristic of crenarchaeal EndAs. Our mutational study revealed that the conserved lysine residue in the loop is important for endonuclease activity. Furthermore, the sequence characteristics of the loops and the positions towards the substrate RNA according to a docking model prompted us to propose that crenarchaea-specific loops and an extra amino acid sequence at the catalytic loop of nanoarchaeal EndA are derived by independent convergent evolution and function for recognizing noncanonical bulge-helix-bulge motif RNAs as substrates.
Organizational Affiliation:
Department of Biomedical Chemistry, University of Tokyo, Tokyo 113-0033, Japan.