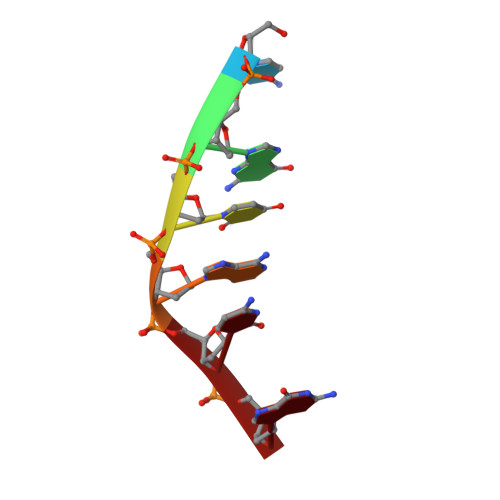
Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
Todd, A.K., Adams, A., Thorpe, J.H., Denny, W.A., Wakelin, L.P.G., Cardin, C.J.(1999) J Med Chem 42: 536-540
- PubMed: 10052960
- DOI: https://doi.org/10.1021/jm980479u
- Primary Citation of Related Structures:
366D, 367D, 452D
Organizational Affiliation:
Chemistry Department, University of Reading, Whiteknights, Reading RG6 6AD, U.K.