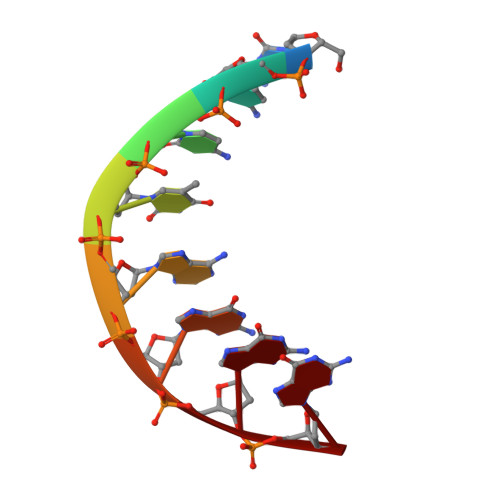
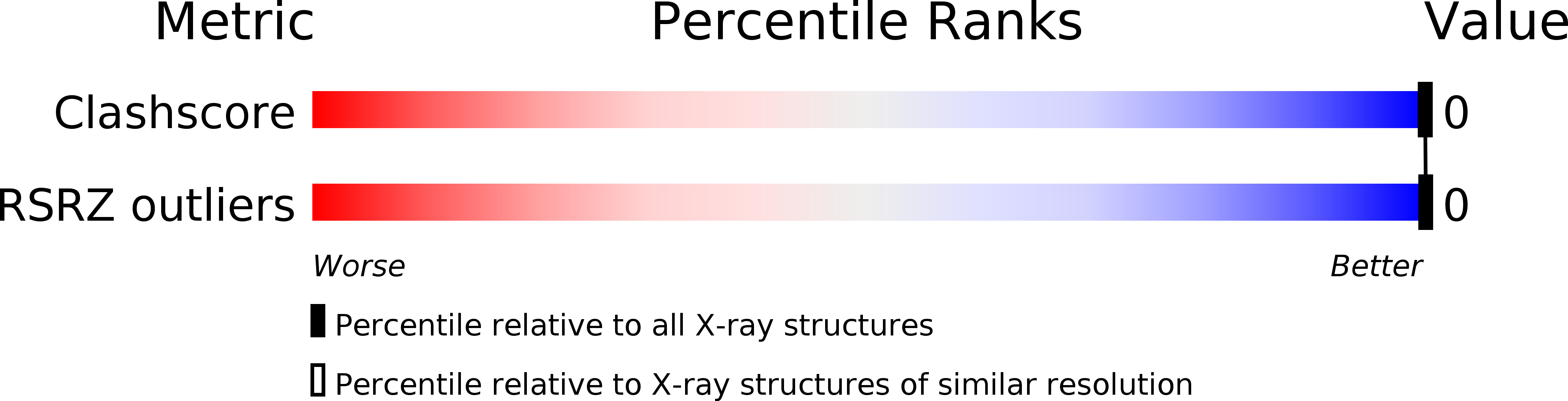Structure of d(CCCTAGGG): comparison with nine isomorphous octamer sequences reveals four distinct patterns of sequence-dependent intermolecular interactions.
Tippin, D.B., Sundaralingam, M.(1996) Acta Crystallogr D Biol Crystallogr 52: 997-1003
- PubMed: 15299609
- DOI: https://doi.org/10.1107/S0907444996005033
- Primary Citation of Related Structures:
317D - PubMed Abstract:
The self-complementary deoxyoctanuceotide d(CCCTAGGG) crystallizes as an A-type double helix in the space group P4(3)2(1)2, a = b = 42.22 and c = 24.90 A, with one strand per asymmetric unit. Using 1533 unique reflections at 1.9 A and I > 2sigma, the structure was solved by molecular placement and refined to a final R value of 16.4%. This structure is isomorphous with nine other tetragonal A-DNA octamers, possessing a central pyrimidine/purine step that is fully extended along the backbone with trans, trans conformations around the C4'-C5' and O5'-P bonds. A structural water, sandwiched between the pi-cloud of the terminal guanine and the N3 atom of G7 in the adjacent duplex, stabilizes an intermolecular base triplet with one hydrogen bond between the terminal cytosine and G6 of an adjacent duplex. Comparative analysis of this structure with the isomorphous A-DNA octamers reveals the importance of base sequence and minor-groove hydration in intermolecular interactions. The minor grooves, which provide both hydrophobic and polar interactions, allow for four patterns of sequence-dependent binding involving interduplex base triplets in which the third base is bonded through a single hydrogen bond. A conserved water molecule appears to be crucial in the stabilization of these intermolecular interactions, which resemble specific recognition motifs found in the crystal structures of the TATA-box/TBP protein complex.
Organizational Affiliation:
Biological Macromolecular Structure, Newman and Wolfram Laboratory of Chemistry, Department of Biochemistry, The Ohio State University, Columbus 43210, USA.