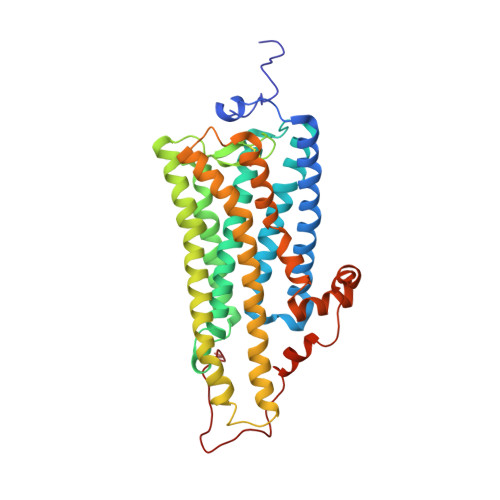
Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region
Shimamura, T., Hiraki, K., Takahashi, N., Hori, T., Ago, H., Masuda, K., Takio, K., Ishiguro, M., Miyano, M.(2008) J Biol Chem 283: 17753-17756
- PubMed: 18463093
- DOI: https://doi.org/10.1074/jbc.C800040200
- Primary Citation of Related Structures:
2ZIY - PubMed Abstract:
G-protein-coupled receptors play a key step in cellular signal transduction cascades by transducing various extracellular signals via G-proteins. Rhodopsin is a prototypical G-protein-coupled receptor involved in the retinal visual signaling cascade. We determined the structure of squid rhodopsin at 3.7A resolution, which transduces signals through the G(q) protein to the phosphoinositol cascade. The structure showed seven transmembrane helices and an amphipathic helix H8 has similar geometry to structures from bovine rhodopsin, coupling to G(t), and human beta(2)-adrenergic receptor, coupling to G(s). Notably, squid rhodopsin contains a well structured cytoplasmic region involved in the interaction with G-proteins, and this region is flexible or disordered in bovine rhodopsin and human beta(2)-adrenergic receptor. The transmembrane helices 5 and 6 are longer and extrude into the cytoplasm. The distal C-terminal tail contains a short hydrophilic alpha-helix CH after the palmitoylated cysteine residues. The residues in the distal C-terminal tail interact with the neighboring residues in the second cytoplasmic loop, the extruded transmembrane helices 5 and 6, and the short helix H8. Additionally, the Tyr-111, Asn-87, and Asn-185 residues are located within hydrogen-bonding distances from the nitrogen atom of the Schiff base.
Organizational Affiliation:
RIKEN SPring-8 Center, Harima Institute, Kouto, Sayo, Hyogo, Japan.