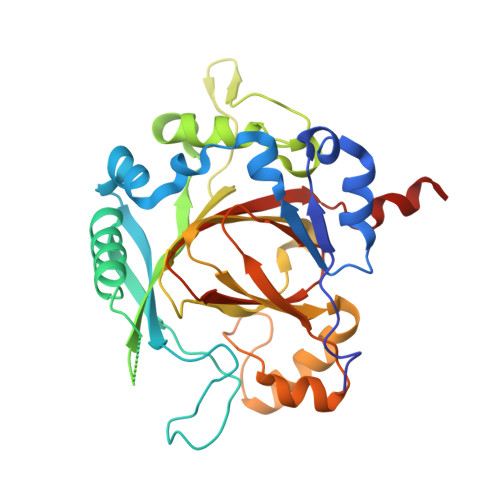
Crystal Structure of the Human Jmjd3 Jumonji Domain
Che, K.H., Yue, W.W., Krojer, T., Muniz, J.R.C., Ng, S.S., Tumber, A., Daniel, M., Burgess-Brown, N., Savitsky, P., von Delft, F., Ugochukwu, E., Filippakopoulos, P., Arrowsmith, C., Weigelt, J., Edwards, A., Bountra, C., Oppermann, U.To be published.