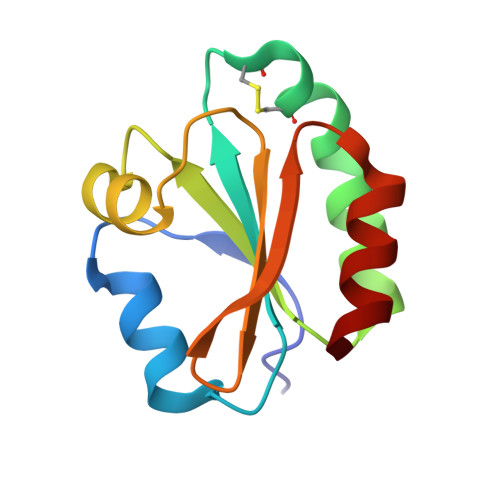
Structural and Functional Characterization of Schistosoma Mansoni Thioredoxin.
Boumis, G., Angelucci, F., Bellelli, A., Brunori, M., Dimastrogiovanni, D., Miele, A.E.(2011) Protein Sci 20: 1069
- PubMed: 21465612
- DOI: https://doi.org/10.1002/pro.634
- Primary Citation of Related Structures:
2XBI, 2XBQ, 2XC2 - PubMed Abstract:
Schistosomiasis, the human parasitosis caused by various species of the blood-fluke Schistosoma, is a debilitating disease affecting 200 million people in tropical areas. The massive administration of the only effective drug, praziquantel, leads to the appearance of less sensitive parasite strains, thus, making urgent the search for new therapeutic approaches and new suitable targets. The thiol-mediated detoxification pathway has been identified as a promising target, being essential during all the parasite developmental stages and sufficiently different from the host counterpart. As a part of a project aimed at the structural characterization of all the proteins involved in this pathway, we describe hereby the high-resolution crystal structure of Schistosoma mansoni Thioredoxin (SmTrx) in three states, namely: the wild-type oxidized adult enzyme and the oxidized and reduced forms of a juvenile isoform, carrying an N-terminal extension. SmTrx shows a typical thioredoxin fold, highly similar to the other components of the superfamily. Although probably unlikely to be a reasonable drug target given its high similarity with the human counterpart, SmTrx completes the characterization of the whole set of thiol-mediated detoxification pathway components. Moreover, it can reduce oxidized glutathione and is one of the few defence proteins expressed in mature eggs and in the hatch fluid, thus confirming an important role in the parasite. We believe its crystal structure may provide clues for the formation of granulomas and the pathogenesis of the chronic disease.
Organizational Affiliation:
Dipartimento di Scienze Biochimiche and Istituto Pasteur-Fondazione Cenci Bolognetti, "Sapienza" University of Rome, P.le Aldo Moro 5, 00185 Rome, Italy.