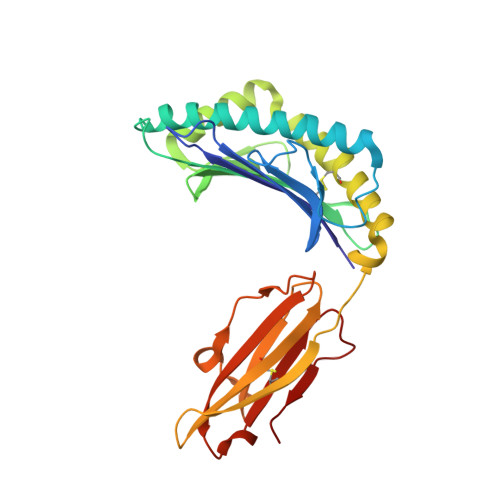
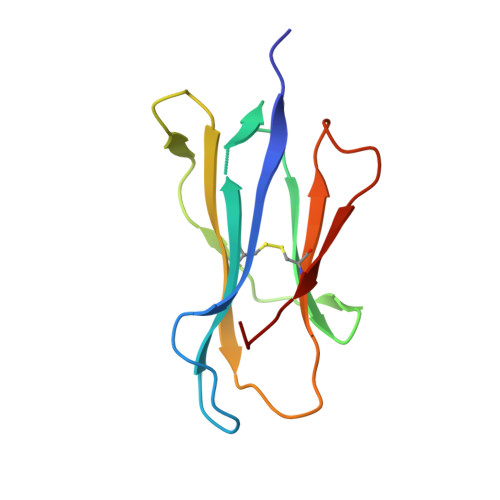
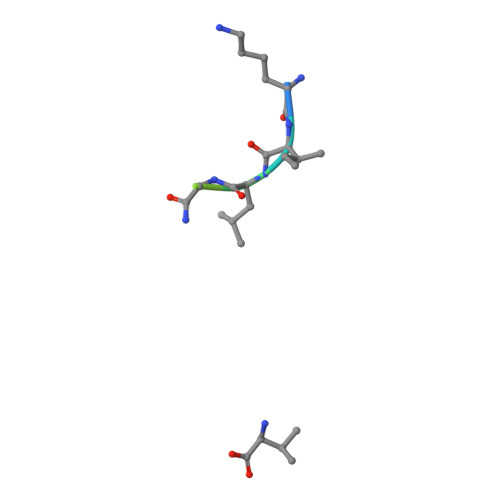
Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
Celie, P.H.N., Toebes, M., Rodenko, B., Ovaa, H., Perrakis, A., Schumacher, T.N.M.(2009) J Am Chem Soc 131: 12298
- PubMed: 19655750
- DOI: https://doi.org/10.1021/ja9037559
- Primary Citation of Related Structures:
2X4N, 2X4O, 2X4R, 2X4S, 2X4U, 2X70 - PubMed Abstract:
High-throughput structure determination of protein-ligand complexes is central in drug development and structural proteomics. To facilitate such high-throughput structure determination we designed an induced replacement strategy. Crystals of a protein complex bound to a photosensitive ligand are exposed to UV light, inducing the departure of the bound ligand, allowing a new ligand to soak in. We exemplify the approach for a class of protein complexes that is especially recalcitrant to high-throughput strategies: the MHC class I proteins. We developed a UV-sensitive, "conditional", peptide ligand whose UV-induced cleavage in the crystals leads to the exchange of the low-affinity lytic fragments for full-length peptides introduced in the crystallant solution. This "in crystallo" exchange is monitored by the loss of seleno-methionine anomalous diffraction signal of the conditional peptide compared to the signal of labeled MHC beta2m subunit. This method has the potential to facilitate high-throughput crystallography in various protein families.
Organizational Affiliation:
Division of Biochemistry, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX, Amsterdam, The Netherlands.