Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Ganesan, R., Eigenbrot, C., Wu, Y., Liang, W.C., Shia, S., Lipari, M.T., Kirchhofer, D.(2009) Structure 17: 1614
- PubMed: 20004165
- DOI: https://doi.org/10.1016/j.str.2009.09.014
- Primary Citation of Related Structures:
2WUB, 2WUC, 3K2U - PubMed Abstract:
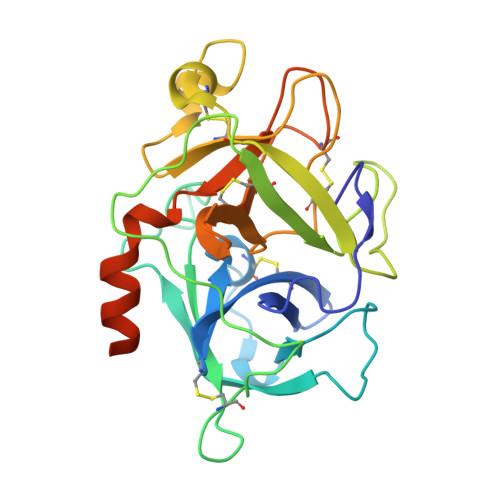
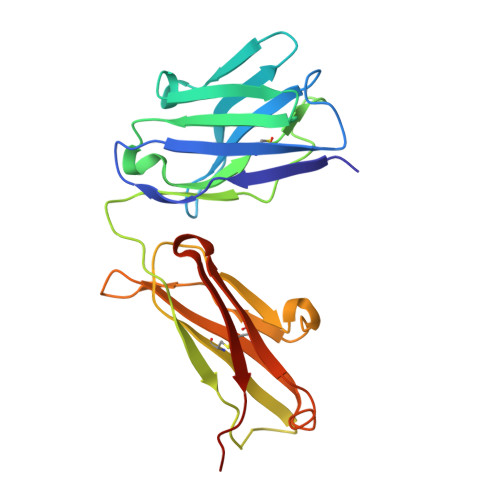
Recent structural studies have outlined the mechanism of protease inhibition by active site-directed antibodies. However, the molecular basis of allosteric inhibition by antibodies has been elusive. Here we report the 2.35 A resolution structure of the trypsin-like serine protease hepatocyte growth factor activator (HGFA) in complex with the allosteric antibody Ab40, a potent inhibitor of HGFA catalytic activity. The antibody binds at the periphery of the substrate binding cleft and imposes a conformational change on the entire 99-loop (chymotrypsinogen numbering). The altered conformation of the 99-loop is incompatible with substrate binding due to the partial collapse of subsite S2 and the reorganization of subsite S4. Remarkably, a single residue deletion of Ab40 abolished inhibition of HGFA activity, commensurate with the reversal of the 99-loop conformation to its "competent" state. The results define an "allosteric switch" mechanism as the basis of protease inhibition by an allosteric antibody.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., South San Francisco, CA 94080, USA.