The Structure of the Membrane Extrinsic Region of Bovine ATP Synthase
Rees, D.M., Leslie, A.G.W., Walker, J.E.(2009) Proc Natl Acad Sci U S A 106: 21597
- PubMed: 19995987
- DOI: https://doi.org/10.1073/pnas.0910365106
- Primary Citation of Related Structures:
2WSS - PubMed Abstract:
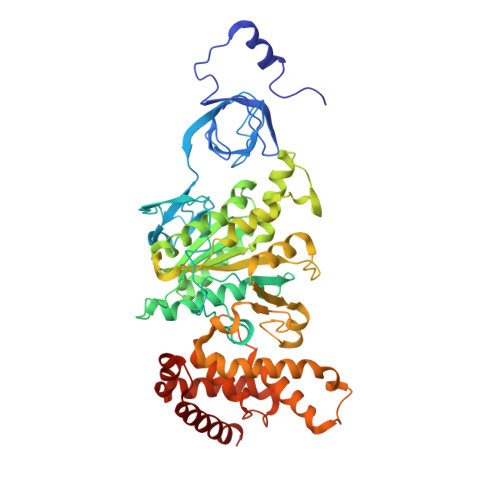
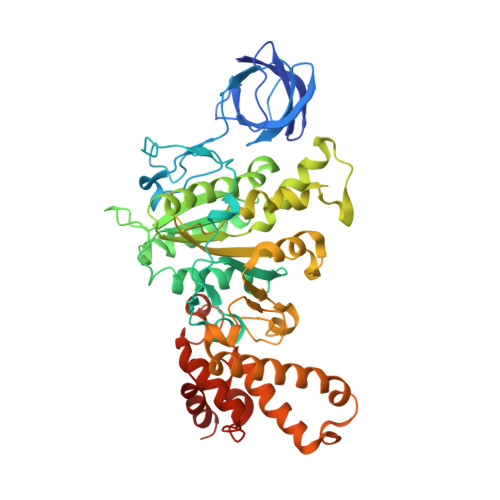
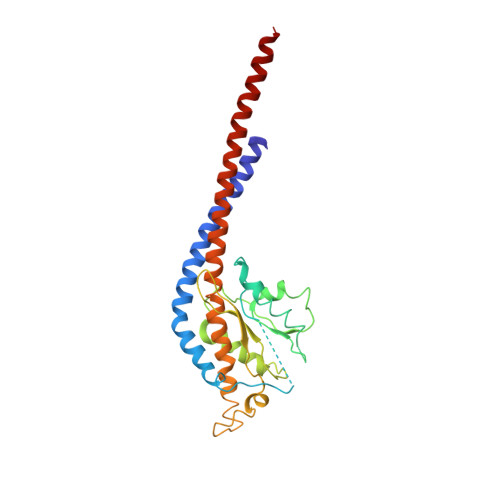
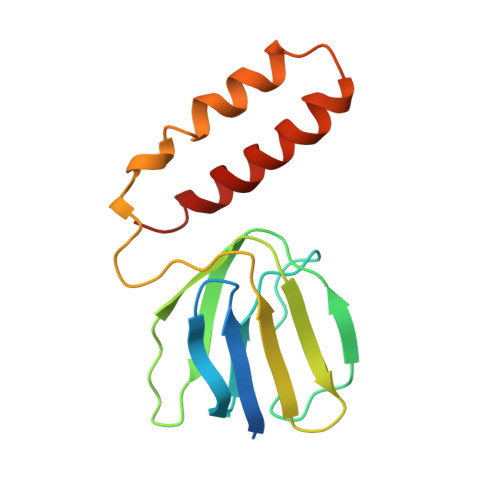
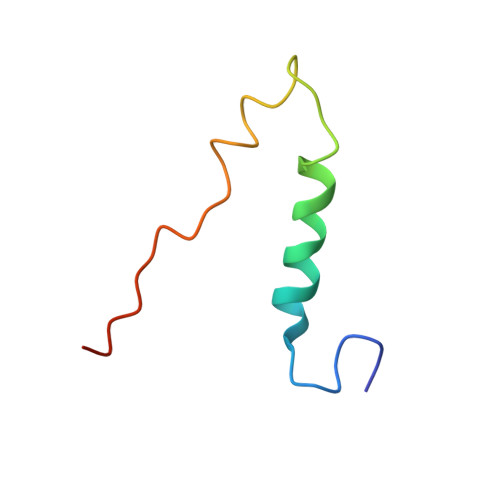
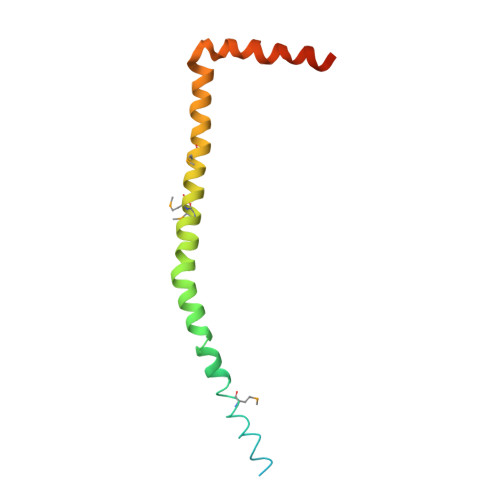
The structure of the complex between bovine mitochondrial F(1)-ATPase and a stator subcomplex has been determined at a resolution of 3.2 A. The resolved region of the stator contains residues 122-207 of subunit b; residues 5-25 and 35-57 of F(6); 3 segments of subunit d from residues 30-40, 65-74, and 85-91; and residues 1-146 and 169-189 of the oligomycin sensitivity conferral protein (OSCP). The stator subcomplex represents its membrane distal part, and its structure has been augmented with an earlier structure of a subcomplex containing residues 79-183, 3-123, and 5-70 of subunits b, d, and F(6), respectively, which extends to the surface of the inner membrane of the mitochondrion. The N-terminal domain of the OSCP links the stator with F(1)-ATPase via alpha-helical interactions with the N-terminal region of subunit alpha(E). Its C-terminal domain makes extensive helix-helix interactions with the C-terminal alpha-helix of subunit b from residues 190-207. Subunit b extends as a continuous 160-A long alpha-helix from residue 188 back to residue 79 near to the surface of the inner mitochondrial membrane. This helix appears to be stiffened by other alpha-helices in subunits d and F(6), but the structure can bend inward toward the F(1) domain around residue 146 of subunit b. The linker region between the 2 domains of the OSCP also appears to be flexible, enabling the stator to adjust its shape as it passes over the changing profile of the F(1) domain during a catalytic cycle. The structure of the membrane extrinsic part of bovine ATP synthase is now complete.
Organizational Affiliation:
Medical Research Council Mitochondrial Biology Unit, Wellcome Trust/Medical Research Council Building, Hills Road, Cambridge CB2 0XY, United Kingdom.