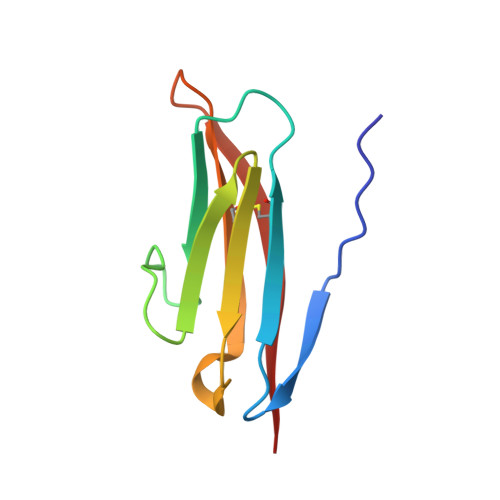
Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
Rasmussen, K.K., Kulahin, N., Kristensen, O., Poulsen, J.C., Sigurskjold, B.W., Kastrup, J.S., Berezin, V., Bock, E., Walmod, P.S., Gajhede, M.(2008) J Mol Biol 382: 1113
- PubMed: 18706912
- DOI: https://doi.org/10.1016/j.jmb.2008.07.084
- Primary Citation of Related Structures:
2VAJ - PubMed Abstract:
The crystal structure of the first immunoglobulin (Ig1) domain of neural cell adhesion molecule 2 (NCAM2/OCAM/RNCAM) is presented at a resolution of 2.7 A. NCAM2 is a member of the immunoglobulin superfamily of cell adhesion molecules (IgCAMs). In the structure, two Ig domains interact by domain swapping, as the two N-terminal beta-strands are interchanged. beta-Strand swapping at the terminal domain is the accepted mechanism of homophilic interactions amongst the cadherins, another class of CAMs, but it has not been observed within the IgCAM superfamily. Gel-filtration chromatography demonstrated the ability of NCAM2 Ig1 to form dimers in solution. Taken together, these observations suggest that beta-strand swapping could have a role in the molecular mechanism of homophilic binding for NCAM2.
Organizational Affiliation:
Protein Laboratory, Department of Neuroscience and Pharmacology, Faculty of Health Sciences, University of Copenhagen, Blegdamsvej 3, DK-2200 Copenhagen, Denmark.