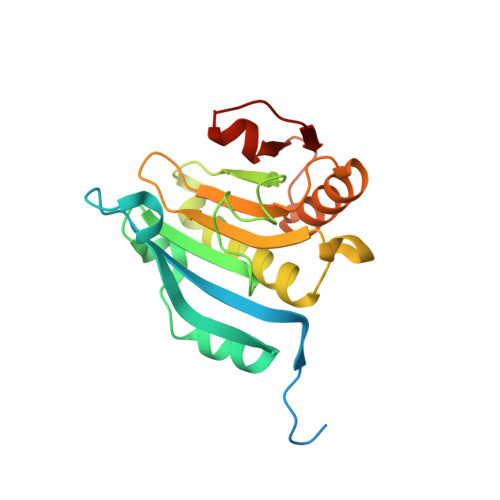
Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
Brown, C.J., Mcnae, I., Fischer, P.M., Walkinshaw, M.D.(2007) J Mol Biol 372: 7
- PubMed: 17631896
- DOI: https://doi.org/10.1016/j.jmb.2007.06.033
- Primary Citation of Related Structures:
2V8W, 2V8X, 2V8Y - PubMed Abstract:
Structural complexes of the eukaryotic translation initiation factor 4E (eIF4E) with a series of N(7)-alkylated guanosine derivative mRNA cap analogue structures have been characterised. Mass spectrometry was used to determine apparent gas-phase equilibrium dissociation constants (K(d)) values of 0.15 microM, 13.6 microM, and 55.7 microM for eIF4E with 7-methyl-GTP (m(7)GTP), GTP, and GMP, respectively. For tight and specific binding to the eIF4E mononucleotide binding site, there seems to be a clear requirement for guanosine derivatives to possess both the delocalised positive charge of the N(7)-methylated guanine system and at least one phosphate group. We show that the N(7)-benzylated monophosphates 7-benzyl-GMP (Bn(7)GMP) and 7-(p-fluorobenzyl)-GMP (FBn(7)GMP) bind eIF4E substantially more tightly than non-N(7)-alkylated guanosine derivatives (K(d) values of 7.0 microM and 2.0 microM, respectively). The eIF4E complex crystal structures with Bn(7)GMP and FBn(7)GMP show that additional favourable contacts of the benzyl groups with eIF4E contribute binding energy that compensates for loss of the beta and gamma-phosphates. The N(7)-benzyl groups pack into a hydrophobic pocket behind the two tryptophan side-chains that are involved in the cation-pi stacking interaction between the cap and the eIF4E mononucleotide binding site. This pocket is formed by an induced fit in which one of the tryptophan residues involved in cap binding flips through 180 degrees relative to structures with N(7)-methylated cap derivatives. This and other observations made here will be useful in the design of new families of eIF4E inhibitors, which may have potential therapeutic applications in cancer.
Organizational Affiliation:
Structural Biochemistry, The University of Edinburgh, Michael Swann Building, King's Buildings, Edinburgh, EH9 3JR, Scotland, UK.