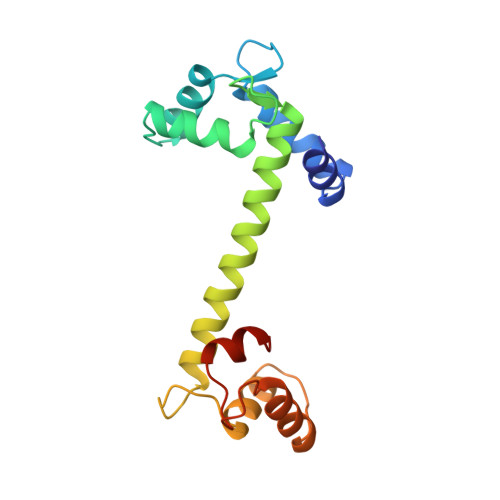
Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Houdusse, A., Love, M.L., Dominguez, R., Grabarek, Z., Cohen, C.(1997) Structure 5: 1695-1711
- PubMed: 9438870
- DOI: https://doi.org/10.1016/s0969-2126(97)00315-8
- Primary Citation of Related Structures:
1TN4, 2TN4 - PubMed Abstract:
In contrast to Ca2+4-bound calmodulin (CaM), which has evolved to bind to many target sequences and thus regulate the function of a variety of enzymes, troponin C (TnC) is a bistable switch which controls contraction in striated muscles. The specific target of TnC is troponin I (TnI), the inhibitory subunit of the troponin complex on the thin filaments of muscle. To date, only the crystal structure of Ca2+2-bound TnC (i.e. in the 'off' state) had been determined, which together with the structure of Ca2+4-bound CaM formed the basis for the so-called 'HMJ' model of the conformational changes in TnC upon Ca2+ binding. NMR spectroscopic studies of Ca2+4-bound TnC (i.e. in the 'on' state) have recently been carried out, but the detailed conformational changes that take place upon switching from the off to the on state have not yet been described. We have determined the crystal structures of two forms of expressed rabbit Ca2+4-bound TnC to 2.0 A resolution. The structures show that the conformation of the N-terminal lobe (N lobe) is similar to that predicted by the HMJ model. Our results also reveal, in detail, the residues involved in binding of Ca2+ in the regulatory N lobe of the molecule. We show that the central helix, which links the N and C lobes of TnC, is better stabilized in the Ca2+2-bound than in the Ca2+4-bound state of the molecule. Comparison of the crystal structures of the off and on states of TnC reveals the specific linkages in the molecule that change in the transition from off to on state upon Ca2+-binding. Small sequence differences are also shown to account for large functional differences between CaM and TnC. The two lobes of TnC are designed to respond to Ca2+-binding quite differently, although the structures with bound Ca2+ are very similar. A small number of differences in the sequences of these two lobes accounts for the fact that the C lobe is stabilized only in the open (Ca2+-bound) state, whereas the N lobe can switch between two stable states. This difference accounts for the Ca2+-dependent and Ca2+-independent interactions of the N and C lobe. The C lobe of TnC is always linked to TnI, whereas the N lobe can maintain its regulatory role - binding strongly to TnI at critical levels of Ca2+ - and in contrast, forming a stable closed conformation in the absence of Ca2+.
Organizational Affiliation:
Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02254-9110, USA.