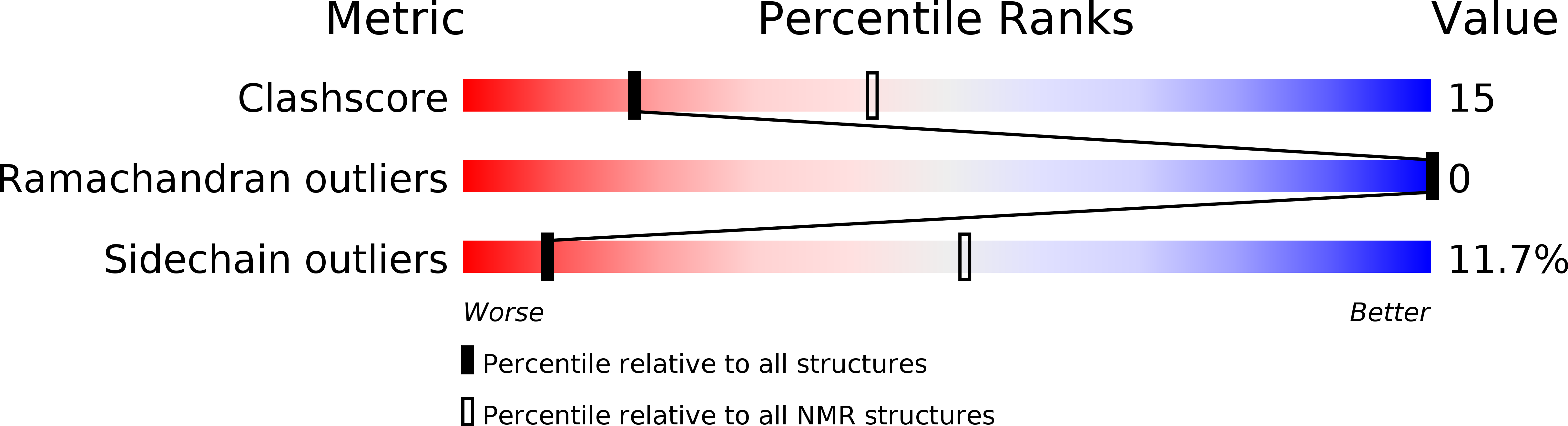Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments.
Umetsu, Y., Tenno, T., Goda, N., Shirakawa, M., Ikegami, T., Hiroaki, H.(2011) Biochim Biophys Acta 1814: 724-730
- PubMed: 21439408
- DOI: https://doi.org/10.1016/j.bbapap.2011.03.009
- Primary Citation of Related Structures:
2RRH, 2RRI - PubMed Abstract:
Vasoactive intestinal peptide (VIP) is a 28-amino acid neuropeptide which belongs to a glucagon/secretin superfamily, the ligand of class II G protein-coupled receptors. Knowledge for the conformation of VIP bound to membrane is important because the receptor activation is initiated by membrane binding of VIP. We have previously observed that VIP-G (glycine-extended VIP) is unstructured in solution, as evidenced by the limited NMR chemical shift dispersion. In this study, we determined the three-dimensional structures of VIP-G in two distinct membrane-mimicking environments. Although these are basically similar structures composed of a disordered N-terminal region and a long α-helix, micelle-bound VIP-G has a curved α-helix. The side chains of residues Phe(6), Tyr(10), Leu(13), and Met(17) found at the concave face form a hydrophobic patch in the micelle-bound state. The structural differences in two distinct membrane-mimicking environments show that the micelle-bound VIP-G localized at the water-micelle boundary with these side chains toward micelle interior. In micelle-bound PACAP-38 (one of the glucagon/secretin superfamily peptide) structure, the identical hydrophobic residues form the micelle-binding interface. This result suggests that these residues play an important role for the membrane binding of VIP and PACAP.
Organizational Affiliation:
Graduate School of Medicine, Kobe University, Chuo-ku, Kobe, Hyogo, Japan.