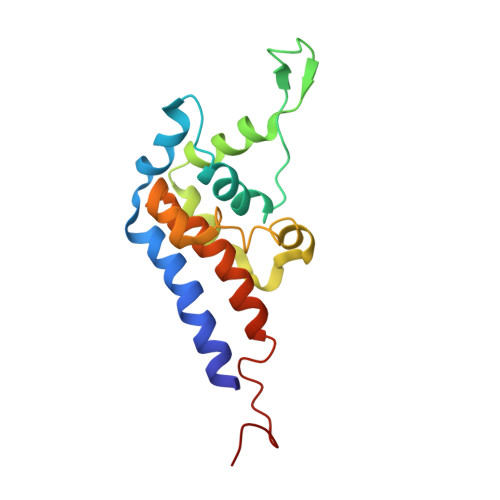
Structure of the microtubule-binding domain of flagellar dynein
Kato, Y.S., Yagi, T., Harris, S.A., Ohki, S.Y., Yura, K., Shimizu, Y., Honda, S., Kamiya, R., Burgess, S.A., Tanokura, M.(2014) Structure 22: 1628-1638
- PubMed: 25450768
- DOI: https://doi.org/10.1016/j.str.2014.08.021
- Primary Citation of Related Structures:
2RR7 - PubMed Abstract:
Flagellar dyneins are essential microtubule motors in eukaryotes, as they drive the beating motions of cilia and flagella. Unlike myosin and kinesin motors, the track binding mechanism of dyneins and the regulation between the strong and weak binding states remain obscure. Here we report the solution structure of the microtubule-binding domain of flagellar dynein-c/DHC9 (dynein-c MTBD). The structure reveals a similar overall helix-rich fold to that of the MTBD of cytoplasmic dynein (cytoplasmic MTBD), but dynein-c MTBD has an additional flap, consisting of an antiparallel b sheet. The flap is positively charged and highly flexible. Despite the structural similarity to cytoplasmic MTBD, dynein-c MTBD shows only a small change in the microtubule- binding affinity depending on the registry change of coiled coil-sliding, whereby lacks the apparent strong binding state. The surface charge distribution of dynein-c MTBD also differs from that of cytoplasmic MTBD, which suggests a difference in the microtubule-binding mechanism.