Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Tsuda, K., Kuwasako, K., Takahashi, M., Someya, T., Inoue, M., Terada, T., Kobayashi, N., Shirouzu, M., Kigawa, T., Tanaka, A., Sugano, S., Guntert, P., Muto, Y., Yokoyama, S.(2009) Nucleic Acids Res 37: 5151-5166
- PubMed: 19553194
- DOI: https://doi.org/10.1093/nar/gkp546
- Primary Citation of Related Structures:
2RQ4, 2RQC - PubMed Abstract:
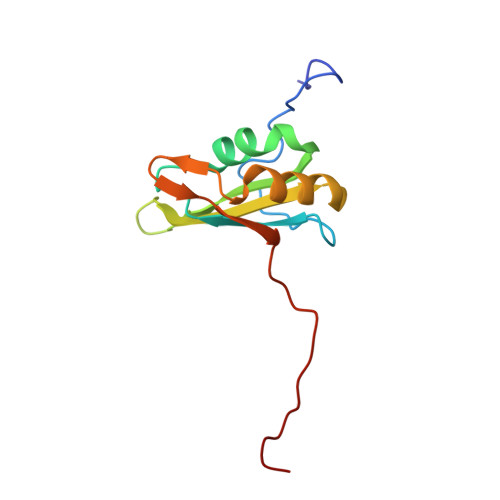
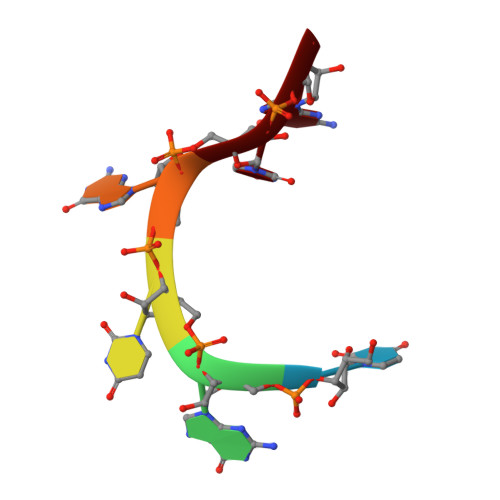
The CUG-binding protein 1 (CUG-BP1) is a member of the CUG-BP1 and ETR-like factors (CELF) family or the Bruno-like family and is involved in the control of splicing, translation and mRNA degradation. Several target RNA sequences of CUG-BP1 have been predicted, such as the CUG triplet repeat, the GU-rich sequences and the AU-rich element of nuclear pre-mRNAs and/or cytoplasmic mRNA. CUG-BP1 has three RNA-recognition motifs (RRMs), among which the third RRM (RRM3) can bind to the target RNAs on its own. In this study, we solved the solution structure of the CUG-BP1 RRM3 by hetero-nuclear NMR spectroscopy. The CUG-BP1 RRM3 exhibited a noncanonical RRM fold, with the four-stranded beta-sheet surface tightly associated with the N-terminal extension. Furthermore, we determined the solution structure of the CUG-BP1 RRM3 in the complex with (UG)(3) RNA, and discovered that the UGU trinucleotide is specifically recognized through extensive stacking interactions and hydrogen bonds within the pocket formed by the beta-sheet surface and the N-terminal extension. This study revealed the unique mechanism that enables the CUG-BP1 RRM3 to discriminate the short RNA segment from other sequences, thus providing the molecular basis for the comprehension of the role of the RRM3s in the CELF/Bruno-like family.
Organizational Affiliation:
RIKEN Systems and Structural Biology Center, Tsurumi, Japan.