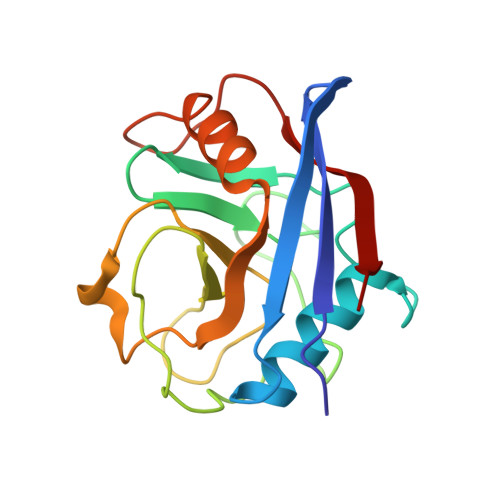
Crystal Structures of Cyclophilin a Complexed with Cyclosporin a and N-Methyl-4-[(E)-2-Butenyl]-4,4-Dimethylthreonine Cyclosporin A.
Ke, H., Mayrose, D., Belshaw, P.J., Alberg, D.G., Schreiber, S.L., Chang, Z.Y., Etzkorn, F.A., Ho, S., Walsh, C.T.(1994) Structure 2: 33
- PubMed: 8075981
- DOI: https://doi.org/10.1016/s0969-2126(00)00006-x
- Primary Citation of Related Structures:
2RMA, 2RMB - PubMed Abstract:
Cyclophilin (CyP) is a ubiquitious intracellular protein that binds the immunosuppressive drug cyclosporin A (CsA). CyP-CsA forms a ternary complex with calcineurin and thereby inhibits T-cell activation. CyP also has enzymatic activity, catalyzing the cis-trans isomerization of peptidyl-prolyl amide bonds.
Organizational Affiliation:
Department of Biochemistry and Biophysics, School of Medicine, University of North Carolina, Chapel HIll 27599.