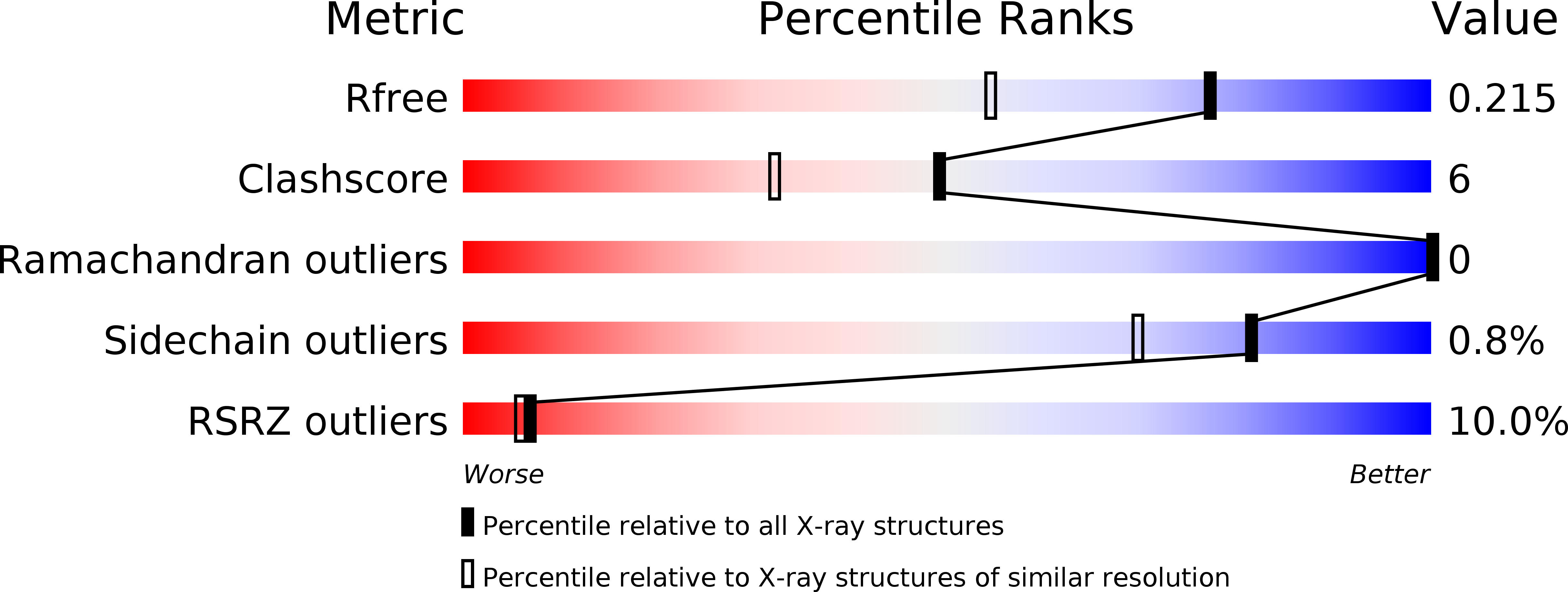
Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Calamini, B., Santarsiero, B.D., Boutin, J.A., Mesecar, A.D.(2008) Biochem J 413: 81-91
- PubMed: 18254726
- DOI: https://doi.org/10.1042/BJ20071373
- Primary Citation of Related Structures:
2QWX, 2QX4, 2QX6, 2QX8, 2QX9 - PubMed Abstract:
Melatonin exerts its biological effects through at least two transmembrane G-protein-coupled receptors, MT1 and MT2, and a lower-affinity cytosolic binding site, designated MT3. MT3 has recently been identified as QR2 (quinone reductase 2) (EC 1.10.99.2) which is of significance since it links the antioxidant effects of melatonin to a mechanism of action. Initially, QR2 was believed to function analogously to QR1 in protecting cells from highly reactive quinones. However, recent studies indicate that QR2 may actually transform certain quinone substrates into more highly reactive compounds capable of causing cellular damage. Therefore it is hypothesized that inhibition of QR2 in certain cases may lead to protection of cells against these highly reactive species. Since melatonin is known to inhibit QR2 activity, but its binding site and mode of inhibition are not known, we determined the mechanism of inhibition of QR2 by melatonin and a series of melatonin and 5-hydroxytryptamine (serotonin) analogues, and we determined the X-ray structures of melatonin and 2-iodomelatonin in complex with QR2 to between 1.5 and 1.8 A (1 A=0.1 nm) resolution. Finally, the thermodynamic binding constants for melatonin and 2-iodomelatonin were determined by ITC (isothermal titration calorimetry). The kinetic results indicate that melatonin is a competitive inhibitor against N-methyldihydronicotinamide (K(i)=7.2 microM) and uncompetitive against menadione (K(i)=92 microM), and the X-ray structures shows that melatonin binds in multiple orientations within the active sites of the QR2 dimer as opposed to an allosteric site. These results provide new insights into the binding mechanisms of melatonin and analogues to QR2.
Organizational Affiliation:
Center for Pharmaceutical Biotechnology and Department of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, The University of Illinois at Chicago, 900 S. Ashland Ave M/C 870, Chicago, IL 60607, USA.