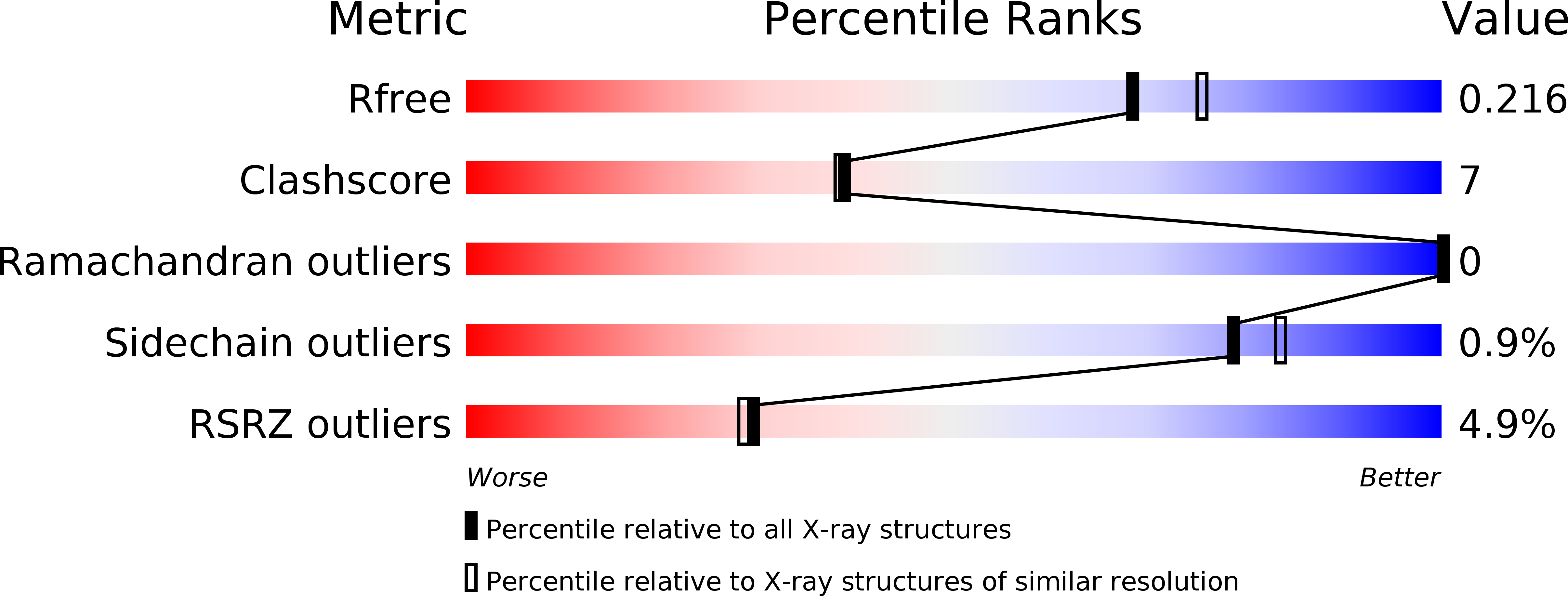
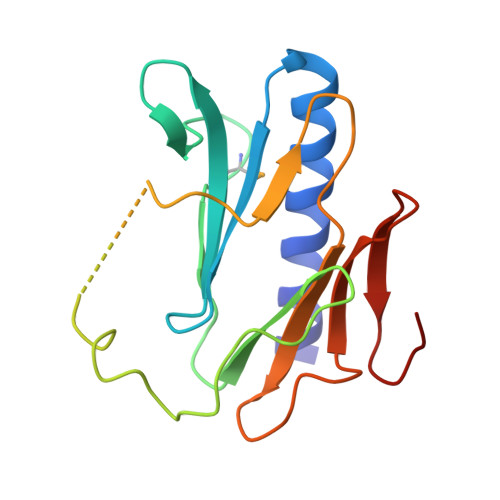
Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae.
Yanez, M.E., Korotkov, K.V., Abendroth, J., Hol, W.G.(2008) J Mol Biol 377: 91-103
- PubMed: 18241884
- DOI: https://doi.org/10.1016/j.jmb.2007.08.041
- Primary Citation of Related Structures:
2QV8 - PubMed Abstract:
Many Gram-negative bacteria use the multi-protein type II secretion system (T2SS) to selectively translocate virulence factors from the periplasmic space into the extracellular environment. In Vibrio cholerae the T2SS is called the extracellular protein secretion (Eps) system,which translocates cholera toxin and several enzymes in their folded state across the outer membrane. Five proteins of the T2SS, the pseudopilins, are thought to assemble into a pseudopilus, which may control the outer membrane pore EpsD, and participate in the active export of proteins in a "piston-like" manner. We report here the 2.0 A resolution crystal structure of an N-terminally truncated variant of EpsH, a minor pseudopilin from Vibrio cholerae. While EpsH maintains an N-terminal alpha-helix and C-terminal beta-sheet consistent with the type 4a pilin fold, structural comparisons reveal major differences between the minor pseudopilin EpsH and the major pseudopilin GspG from Klebsiella oxytoca: EpsH contains a large beta-sheet in the variable domain, where GspG contains an alpha-helix. Most importantly, EpsH contains at its surface a hydrophobic crevice between its variable and conserved beta-sheets, wherein a majority of the conserved residues within the EpsH family are clustered. In a tentative model of a T2SS pseudopilus with EpsH at its tip, the conserved crevice faces away from the helix axis. This conserved surface region may be critical for interacting with other proteins from the T2SS machinery.
Organizational Affiliation:
Department of Biochemistry, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, WA 98195, USA.