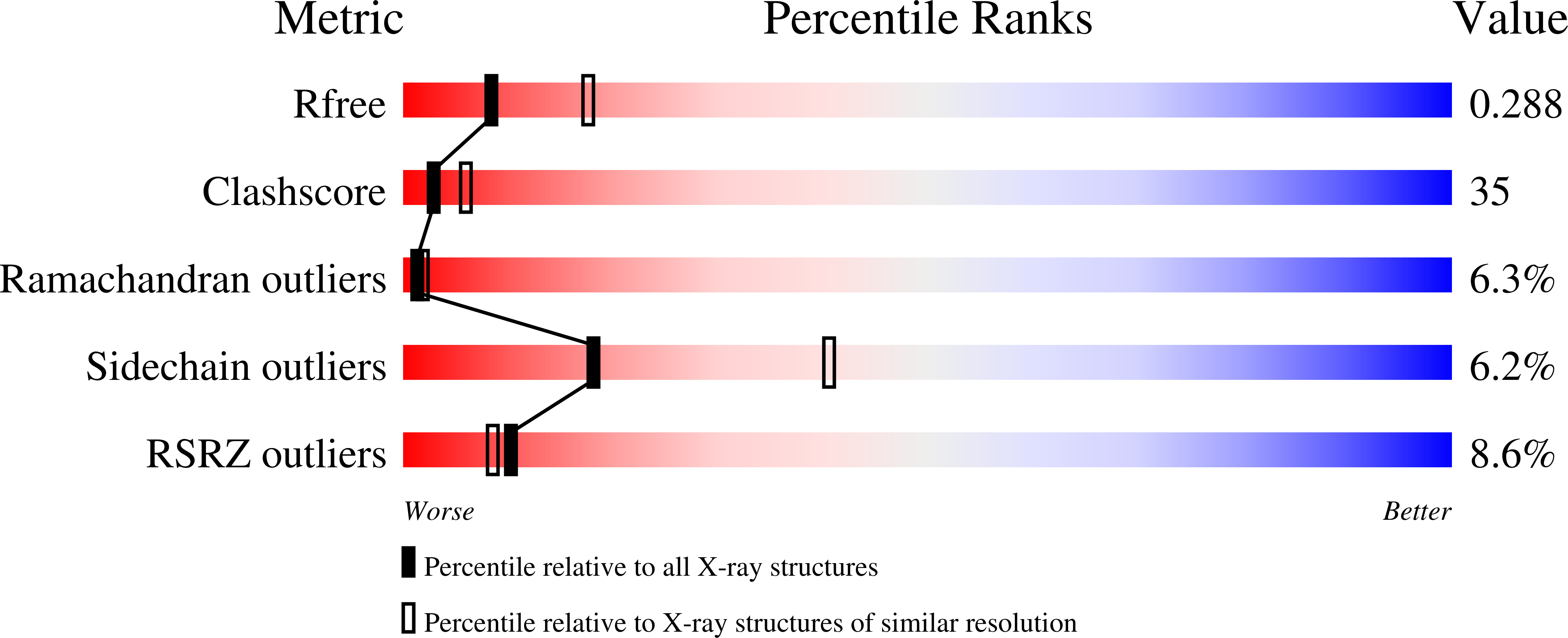
Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation.
Pattanayek, R., Williams, D.R., Pattanayek, S., Mori, T., Johnson, C.H., Stewart, P.L., Egli, M.(2008) EMBO J 27: 1767-1778
- PubMed: 18497745
- DOI: https://doi.org/10.1038/emboj.2008.104
- Primary Citation of Related Structures:
2QKE - PubMed Abstract:
The circadian clock of the cyanobacterium Synechococcus elongatus can be reconstituted in vitro by the KaiA, KaiB and KaiC proteins in the presence of ATP. The principal clock component, KaiC, undergoes regular cycles between hyper- and hypo-phosphorylated states with a period of ca. 24 h that is temperature compensated. KaiA enhances KaiC phosphorylation and this enhancement is antagonized by KaiB. Throughout the cycle Kai proteins interact in a dynamic manner to form complexes of different composition. We present a three-dimensional model of the S. elongatus KaiB-KaiC complex based on X-ray crystallography, negative-stain and cryo-electron microscopy, native gel electrophoresis and modelling techniques. We provide experimental evidence that KaiB dimers interact with KaiC from the same side as KaiA and for a conformational rearrangement of the C-terminal regions of KaiC subunits. The enlarged central channel and thus KaiC subunit separation in the C-terminal ring of the hexamer is consistent with KaiC subunit exchange during the dephosphorylation phase. The proposed binding mode of KaiB explains the observation of simultaneous binding of KaiA and KaiB to KaiC, and provides insight into the mechanism of KaiB's antagonism of KaiA.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, TN 37232, USA.