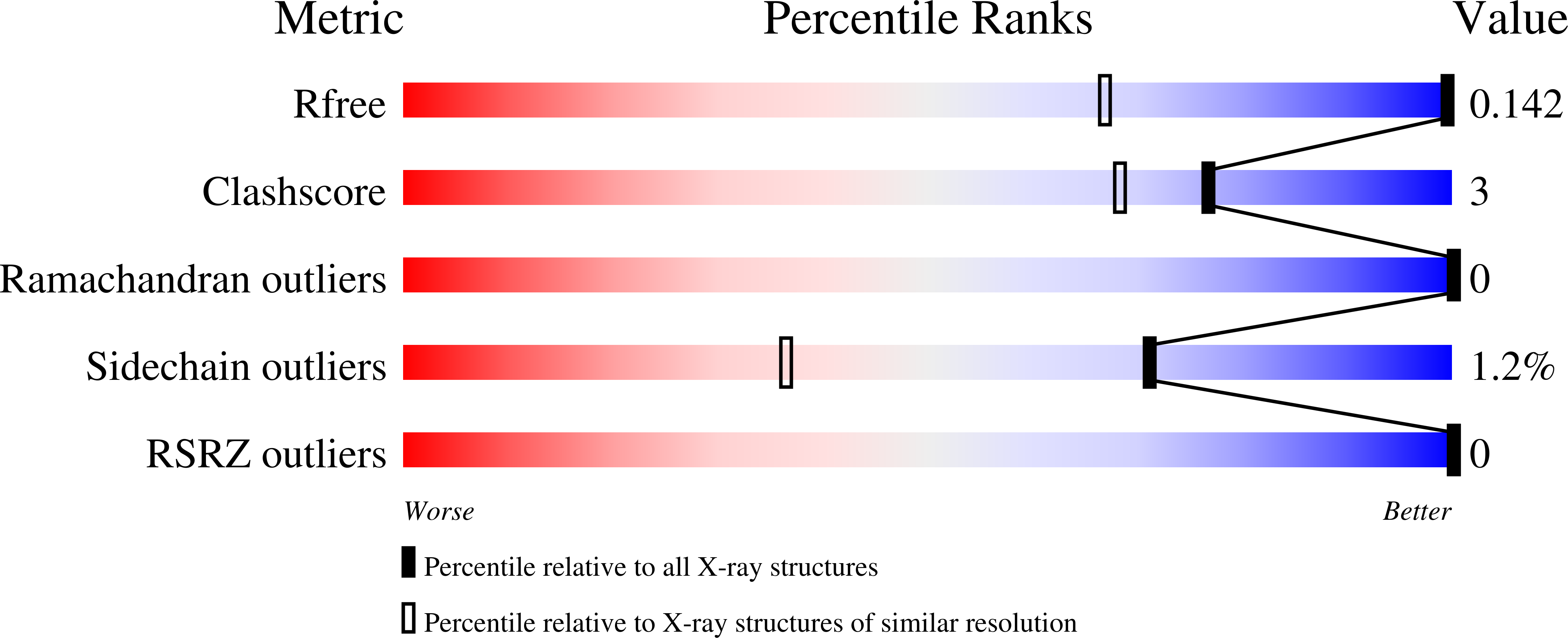
A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Ma, J.K., Wang, Y., Carrell, C.J., Mathews, F.S., Davidson, V.L.(2007) Biochemistry 46: 11137-11146
- PubMed: 17824674
- DOI: https://doi.org/10.1021/bi7012307
- Primary Citation of Related Structures:
2QDV, 2QDW - PubMed Abstract:
Amicyanin is a type 1 copper protein that is the natural electron acceptor for the quinoprotein methylamine dehydrogenase (MADH). A P52G amicyanin mutation increased the Kd for complex formation and caused the normally true electron transfer (ET) reaction from O-quinol MADH to amicyanin to become a gated ET reaction (Ma, J. K., Carrell, C. J., Mathews, F. S., and Davidson, V. L. (2006) Biochemistry 45, 8284-8293). One consequence of the P52G mutation was to reposition the side chain of Met51, which is present at the MADH-amicyanin interface. To examine the precise role of Met51 in this interprotein ET reaction, Met51 was converted to Ala, Lys, and Leu. The Kd for complex formation of M51A amicyanin was unchanged but the experimentally determined electronic coupling increased from 12 cm-1 to 142 cm-1, and the reorganization energy increased from 2.3 to 3.1 eV. The rate and salt dependence of the proton transfer-gated ET reaction from N-quinol MADH to amicyanin is also changed by the M51A mutation. These changes in ET parameters and rates for the reactions with M51A amicyanin were similar to those caused by the P52G mutation and indicated that the ET reaction had become gated by a similar process, most likely a conformational rearrangement of the protein ET complex. The results of the M51K and M51L mutations also have consequences on the kinetic mechanism of regulation of the interprotein ET with effects that are intermediate between what is observed for the reaction of the native amicyanin and M51A amicyanin. These data indicate that the loss of the interactions involving Pro52 were primarily responsible for the change in Kd for P52G amicyanin, while the interactions involving the Met51 side chain are entirely responsible for the change in ET parameters and conversion of the true ET reaction of native amicyanin into a conformationally gated ET reaction.
Organizational Affiliation:
Department of Biochemistry, The University of Mississippi Medical Center, Jackson, Mississippi 39216-4505, USA.