Plant hemoglobins: a molecular fossil record for the evolution of oxygen transport
Hoy, J.A., Robinson, H., Trent III, J.T., Kakar, S., Smagghe, B.J., Hargrove, M.S.(2007) J Mol Biol 371: 168-179
- PubMed: 17560601
- DOI: https://doi.org/10.1016/j.jmb.2007.05.029
- Primary Citation of Related Structures:
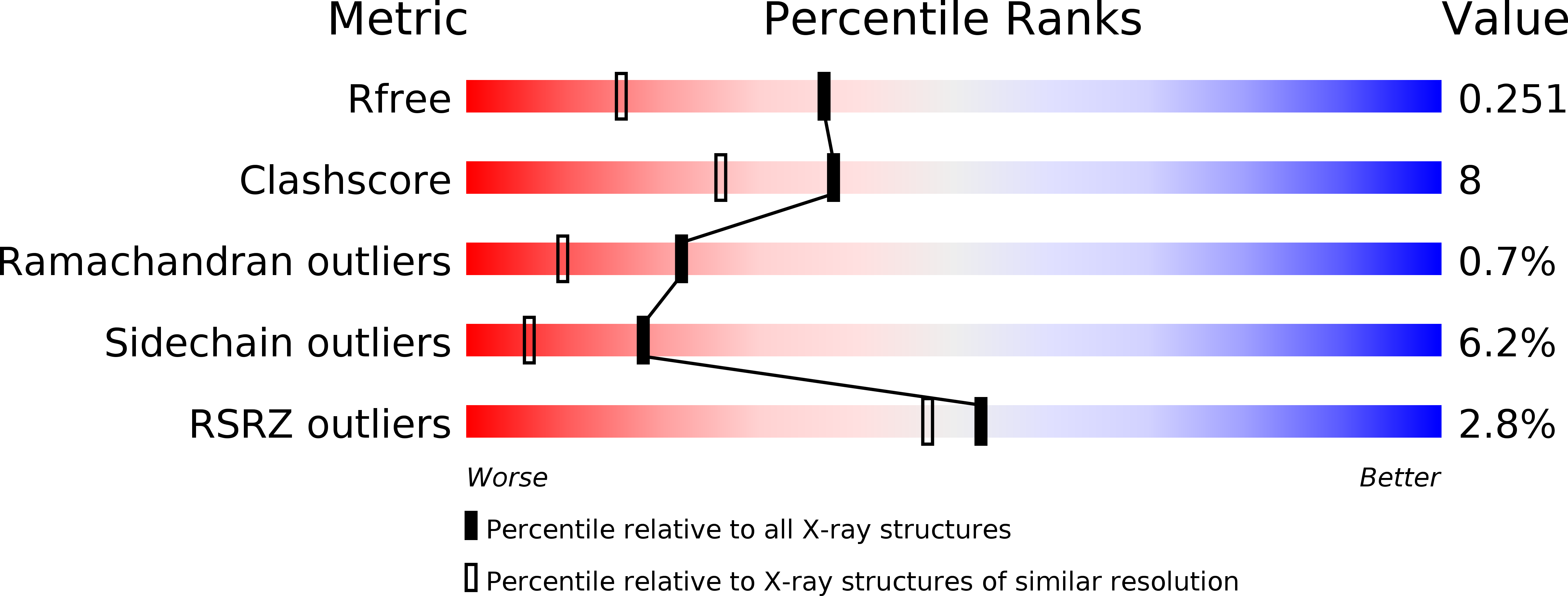
2OIF - PubMed Abstract:
The evolution of oxygen transport hemoglobins occurred on at least two independent occasions. The earliest event led to myoglobin and red blood cell hemoglobin in animals. In plants, oxygen transport "leghemoglobins" evolved much more recently. In both events, pentacoordinate heme sites capable of inert oxygen transfer evolved from hexacoordinate hemoglobins that have unrelated functions. High sequence homology between hexacoordinate and pentacoordinate hemoglobins in plants has poised them for potential structural analysis leading to a molecular understanding of this important evolutionary event. However, the lack of a plant hexacoordinate hemoglobin structure in the exogenously ligand-bound form has prevented such comparison. Here we report the crystal structure of the cyanide-bound hexacoordinate hemoglobin from barley. This presents the first opportunity to examine conformational changes in plant hexacoordinate hemoglobins upon exogenous ligand binding, and reveals structural mechanisms for stabilizing the high-energy pentacoordinate heme conformation critical to the evolution of reversible oxygen binding hemoglobins.
Organizational Affiliation:
Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, IA 50011, USA.