(1)H, (13)C and (15)N resonance assignments and secondary structure analysis of translation initiation factor 1 from Pseudomonas aeruginosa.
Bernal, A., Hu, Y., Palmer, S.O., Silva, A., Bullard, J., Zhang, Y.(2016) Biomol NMR Assign 10: 249-252
- PubMed: 26983940
- DOI: https://doi.org/10.1007/s12104-016-9678-7
- Primary Citation of Related Structures:
2N78 - PubMed Abstract:
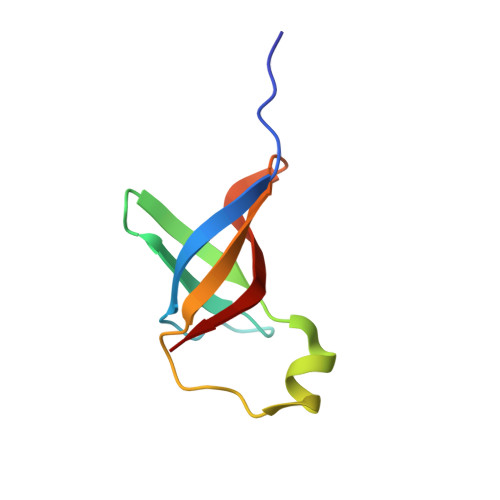
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen and a primary cause of infection in humans. P. aeruginosa can acquire resistance against multiple groups of antimicrobial agents, including β-lactams, aminoglycosides and fluoroquinolones, and multidrug resistance is increasing in this organism which makes treatment of the infections difficult and expensive. This has led to the unmet need for discovery of new compounds distinctly different from present antimicrobials. Protein synthesis is an essential metabolic process and a validated target for the development of new antibiotics. Translation initiation factor 1 from P. aeruginosa (Pa-IF1) is the smallest of the three initiation factors that acts to establish the 30S initiation complex to initiate translation during protein biosynthesis, and its structure is unknown. Here we report the (1)H, (13)C and (15)N chemical shift assignments of Pa-IF1 as the basis for NMR structure determination and interaction studies. Secondary structure analyses deduced from the NMR chemical shift data have identified five β-strands with an unusually extended β-strand at the C-terminal end of the protein and one short α-helix arranged in the sequential order β1-β2-β3-α1-β4-β5. This is further supported by (15)N-{(1)H} hetero NOEs. These secondary structure elements suggest the Pa-IF1 adopts the typical β-barrel structure and is composed of an oligomer-binding motif.
Organizational Affiliation:
Department of Chemistry, The University of Texas Rio Grande Valley, Edinburg, TX, USA.