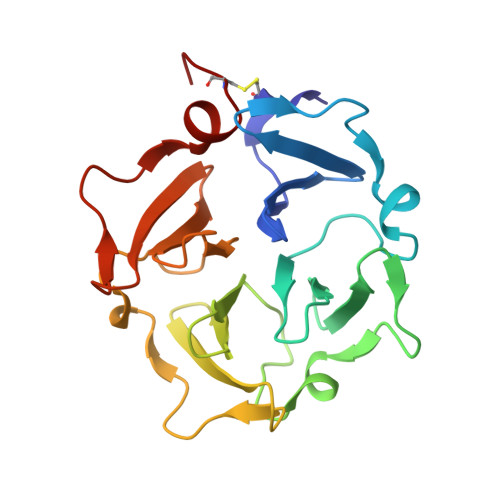
Transient collagen triple helix binding to a key metalloproteinase in invasion and development.
Zhao, Y., Marcink, T.C., Sanganna Gari, R.R., Marsh, B.P., King, G.M., Stawikowska, R., Fields, G.B., Van Doren, S.R.(2015) Structure 23: 257-269
- PubMed: 25651059
- DOI: https://doi.org/10.1016/j.str.2014.11.021
- Primary Citation of Related Structures:
2MQS - PubMed Abstract:
Skeletal development and invasion by tumor cells depends on proteolysis of collagen by the pericellular metalloproteinase MT1-MMP. Its hemopexin-like (HPX) domain binds to collagen substrates to facilitate their digestion. Spin labeling and paramagnetic nuclear magnetic resonance (NMR) detection have revealed how the HPX domain docks to collagen I-derived triple helix. Mutations impairing triple-helical peptidase activity corroborate the interface. Saturation transfer difference NMR suggests rotational averaging around the longitudinal axis of the triple-helical peptide. Part of the interface emerges as unique and potentially targetable for selective inhibition. The triple helix crosses the junction of blades I and II at a 45° angle to the symmetry axis of the HPX domain, placing the scissile Gly∼Ile bond near the HPX domain and shifted ∼25 Å from MMP-1 complexes. This raises the question of the MT1-MMP catalytic domain folding over the triple helix during catalysis, a possibility accommodated by the flexibility between domains suggested by atomic force microscopy images.
Organizational Affiliation:
Department of Biochemistry, University of Missouri, 117 Schweitzer Hall, Columbia, MO 65211, USA.