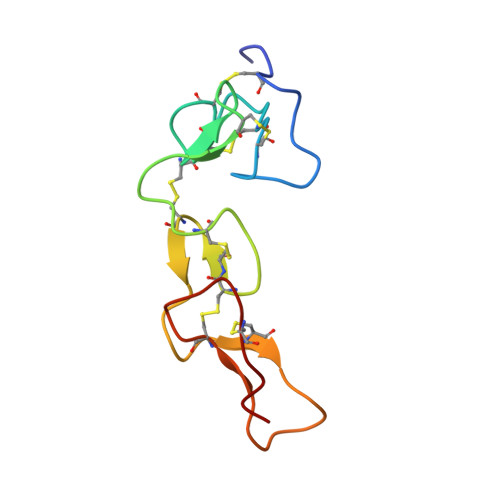
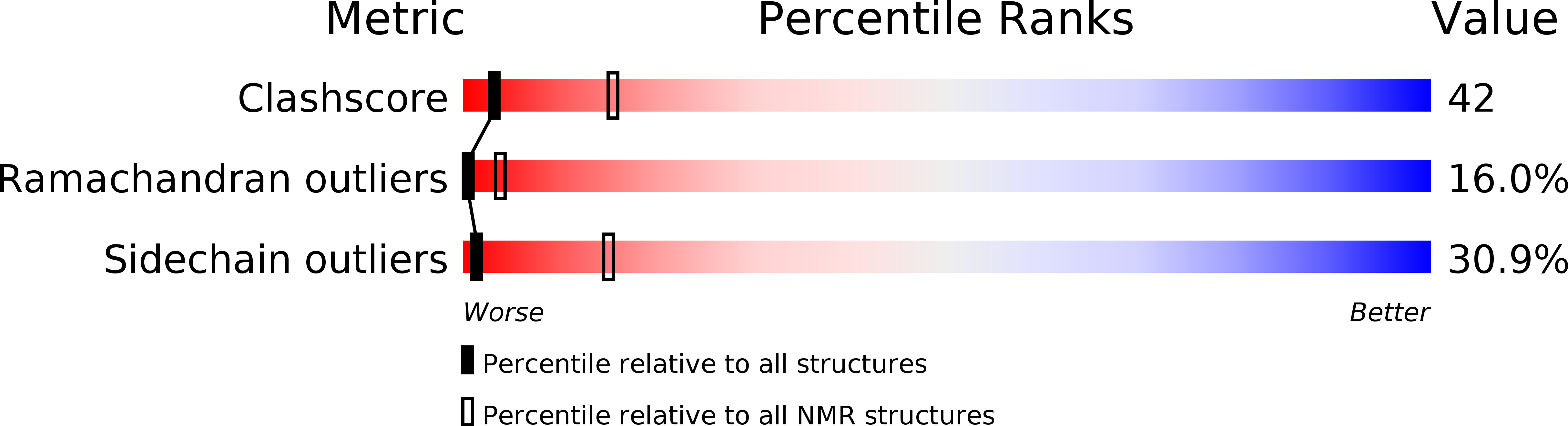NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Carbajo, R.J., Sanz, L., Perez, A., Calvete, J.J.(2015) FEBS J 282: 341-360
- PubMed: 25363287
- DOI: https://doi.org/10.1111/febs.13138
- Primary Citation of Related Structures:
2MOP, 2MP5 - PubMed Abstract:
Extant disintegrins, as found in the venoms of Viperidae and Crotalidae snakes (vipers and rattlesnakes, represent a family of polypeptides that block the function of β1 and β3 integrin receptors, both potently and with a high degree of selectivity. This toxin family owes its origin to the neofunctionalization of the extracellular region of an ADAM (a disintegrin and metalloprotease) molecule recruited into the snake venom gland proteome in the Jurassic. The evolutionary structural diversification of the disintegrin scaffold, from the ancestral long disintegrins to the more recently evolved medium-sized, dimeric and short disintegrins, involved the stepwise loss of pairs of class-specific disulfide linkages and the processing of the N-terminal region. NMR and crystal structures of medium-sized, dimeric and short disintegrins have been solved. However, the structure of a long disintegrin remained unknown. The present study reports the NMR solution structures of two disulfide bond conformers of the long disintegrin bitistatin from the African puff adder Bitis arietans. The findings provide insight into how a structural domain of the extracellular region of an ADAM molecule, recruited into and selectively expressed in the snake venom gland proteome as a PIII metalloprotease in the Jurassic, has subsequently been tranformed into a family of integrin receptor antagonists.
Organizational Affiliation:
Laboratory of Structural Biochemistry, Centro de Investigación Príncipe Felipe, Valencia, Spain.